Advanced Models of Parkinson's Disease
February 22, 2018, at 12:00 PM ETAbstract
Optimized cell lines are essential for modeling neurodegenerative diseases such as Parkinson’s disease, screening novel therapeutics for preclinical studies, and testing the neurotoxicity of environmental compounds. Neural progenitor cells (NPCs) derived from induced pluripotent stem cells (iPSCs) are excellent in vitro models as they can be induced to differentiate down all three neural lineages. ATCC has recently added a line of NPCs developed from a donor with Parkinson’s disease to its collection of neurological research tools. This webinar will describe how ATCC NPCs can be differentiated into three neural fates and used in toxicological studies, focusing on the performance of the Parkinson’s disease-derived NPCs.
Key Points
- ATCC has a wide range of whole-cell models of Parkinson’s disease
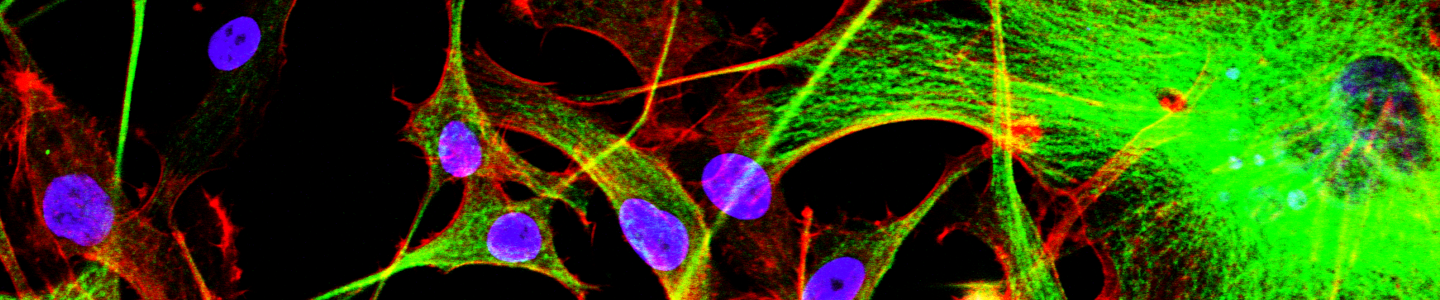
- NPCs cultured in ATCC Dopaminergic Neuron Differentiation Media express tyrosine hydroxylase and TuJ1
- Parkinson’s disease patient-derived NPCs can be induced to differentiate into all three major neural lineages
- Parkinson’s disease patient-derived NPCs can be used to screen compounds for toxicity
Presenter
Brian Shapiro, PhD
Marketing Segment Manager, Oncology, ATCC
Brian A Shapiro, PhD, works to communicate the scientific breakthroughs of ATCC’s product development laboratories to the biomedical research community. Brian is the Executive Producer of ATCC's Podcast, Behind the Biology. Previously, he worked at Virginia Commonwealth University, where he investigated the role of pre-mRNA splicing in the multi-drug resistance of lung cancer. Dr. Shapiro attended the Medical College of Georgia, where his research focused on adrenal physiology as well as diseases of the epidermis.
Questions and Answers
Are there passaging recommendations for NPCs during tri-lineage differentiation?
It is OK to split cells during astrocyte and oligodendrocyte differentiation when NPCs become confluent. However, passaged NPCs will not survive well during dopaminergic neuron differentiation. Over-confluence of NPCs will not affect the outcome of dopaminergic neuron differentiation.
Can you explain why you are getting different neuronal population when you claim your media is dopaminergic differentiation media?
The majority of the growth factors in our differentiation medium are common to generate all neuronal cell types; the big difference is the concentration needed to make a majority of a specific neuron type. By tweaking the concentration of growth factors we were able to increase the number of dopaminergic neurons relative to other neuron types.
You mentioned the addition of CHIR-99021 to the media for differentiating the Parkinson’s disease NPCs into dopaminergic neurons. Where did you get the compound from, and what concentration is it used?
For differentiating ACS-5001 NPCs to dopaminergic neurons, we must add CHIR-99021 at a final concentration of 5 µM (Stemgent cat. 04-0004-10) to the dopaminergic differentiating media (ATCC ACS-3004). This component can be added in an aliquot of dopaminergic media and store it at 4 C for a week.
How many passages can Neural Progenitor Cells Derived from ATCC-DYS0530 Parkinson's Disease (ATCC ACS-5001) undergo?
ATCC NPCs can be proliferated at least 15 PDLs and passaged over 20 times. Some of the results shown in the webinar were using cells from passage 10.
What dissociation reagent is recommended for the passaging of NPCs?
ACS-5001 NPCs require undiluted Accutase for passaging that differs from the protocol for normal NPCs which uses Accutase diluted 1:1 with DPBS.
What is the optimal cell confluence when passaging NPCs?
Unlike other primary cells, passaging NPCs near 100% confluence doesn’t affect cell performance. In fact, it is highly recommended to passage NPCs when they reach ~95% confluence. Do not split NPCs when they are < 85% confluence, which will inhibit cell proliferation.
