Britany Tang, BS;1 Michael Geimer, BS;1 Sung-Oui Suh, PhD;1 Trudy Correia, MS;1 Solmaz Eskandarinezhad, BASc;1,2 Holly Ann Asbury, BS;1 Ann Wasko, BS;1,2 Brian Chase, MS;1 James Budnick, PhD;1 Sujatha Rashid, PhD;1,2 Katalin Kiss, PhD;1 Rebecca Bradford, MBA, MS;1,2 Leka Papazisi, DVM, PhD;1 Victoria Knight-Connoni, PhD1
1ATCC, Manassas, VA 20110
2BEI Resources, Manassas, VA 20110
Background and Introduction
Materials and Methods
Results
Conclusion
References
Abstract
Mpox is a zoonotic viral disease caused by the human monkeypox virus (hMPXV), which is endemic to several countries in central and western Africa and has been increasingly appearing in urban areas. hMPXV causes a smallpox-like disease in humans. Therefore, the rapid and accurate detection of this disease during the early stages of infection is essential for containment and timely and proper treatment. PCR-based methods have been shown to be rapid and highly sensitive, but like any other analytical tests, the development and validation of the PCR assays are dependent on the availability of high-quality reference materials. To address this need, ATCC designed and developed a quantitative synthetic molecular standard for hMPXV, which can serve as a safe and reliable positive control material when conducting diagnostics assays for hMPXV.
Download a PDF of this application note
Download nowBackground and Introduction
Mpox is a zoonotic viral disease caused by hMPXV, which is endemic to several countries in central and western Africa and has been increasingly appearing in urban areas. Belonging to the genus Orthopoxvirus and the family Poxviridae, hMPXV causes a smallpox-like disease in humans. Symptoms typically last between 2 to 4 weeks and include fever, intense headache, muscle aches, back pain, low energy, swollen lymph nodes, and a skin rash or lesions. Therefore, the rapid and accurate diagnosis of this disease during the early stages of infection is essential for timely and proper treatment.
While culture-based approaches can be used to detect hMPXV, they are typically time-consuming, labor-intensive, and require BSL-3 facilities. PCR-based methods provide a highly sensitive and rapid alternative screening approach. However, the development and validation of these assays are dependent on the availability of high-quality reference materials. To address this need, ATCC designed and developed a quantitative synthetic molecular standard for hMPXV that contains biomarkers for Clade I (Congo Basin) and Clade II (West African).4 Here, we used a proprietary strategy to incorporate genes typically targeted in various assays for viral detection and identification. As a proof-of-concept, the hMPXV standard was quantified using validated Droplet Digital™ PCR (ddPCR™; Bio-Rad) via published quantitative PCR (qPCR) assays.1,4 It was further tested using the Centers for Disease Control and Prevention’s (CDC) non-variola Orthopoxvirus and generic Monkeypox virus assays.2,3 Our results indicate that the designed synthetic DNA construct is compatible with all assays tested.
Materials and Methods
Quantitative Synthetic DNA
Using a proprietary method, we designed a synthetic DNA construct for hMPXV (Quantitative Synthetic Monkeypox virus DNA; ATCC VR-3270SD). This standard comprises fragments from the following genomic regions: J2L, D14L, F3L, F8L, A27L, A29L, B6R, B7R, and N3R. Following construction, the standard was authenticated via next-generation sequencing and then quantified via ddPCR.
qPCR Assay
qPCR assays were performed using the CFX96™ Real-Time PCR Detection System (Bio-Rad) according to the manufacturer’s instructions. Details for individual assays are described in Figures 1, 3, 4, and 5 using ATCC VR-3270SD as the standard.
ddPCR Assay
ddPCR assays were performed according to the manufacturer’s instructions using the QX200™ droplet reader and QuantaSoft™ software 1.7.4.0917 (Bio-Rad) for droplet generation and data analysis. ddPCR was adapted from an existing protocol.1
Results
We conducted functional testing of the synthetic material via four published PCR assays (Figures 1-5).
| A | B |
 |
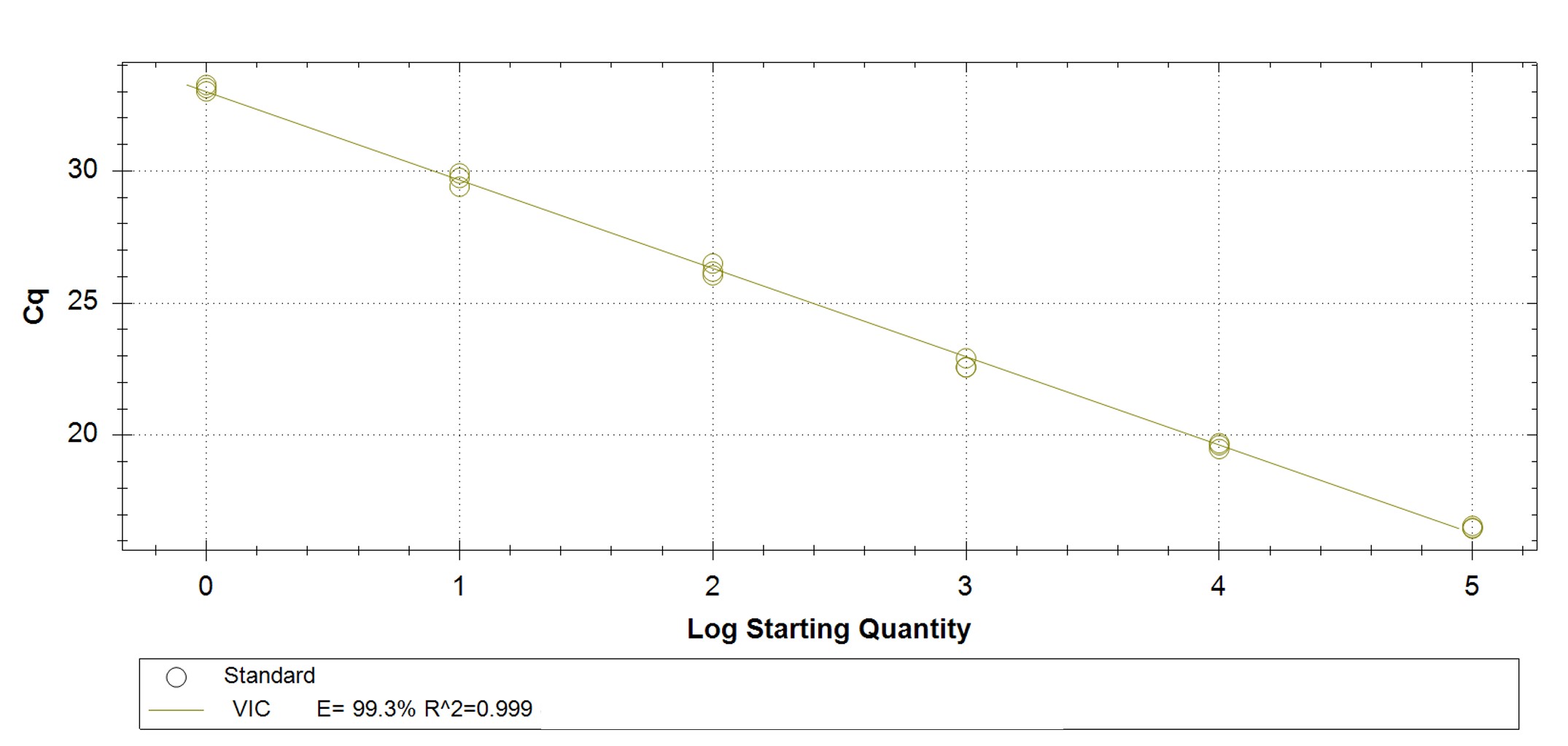 |
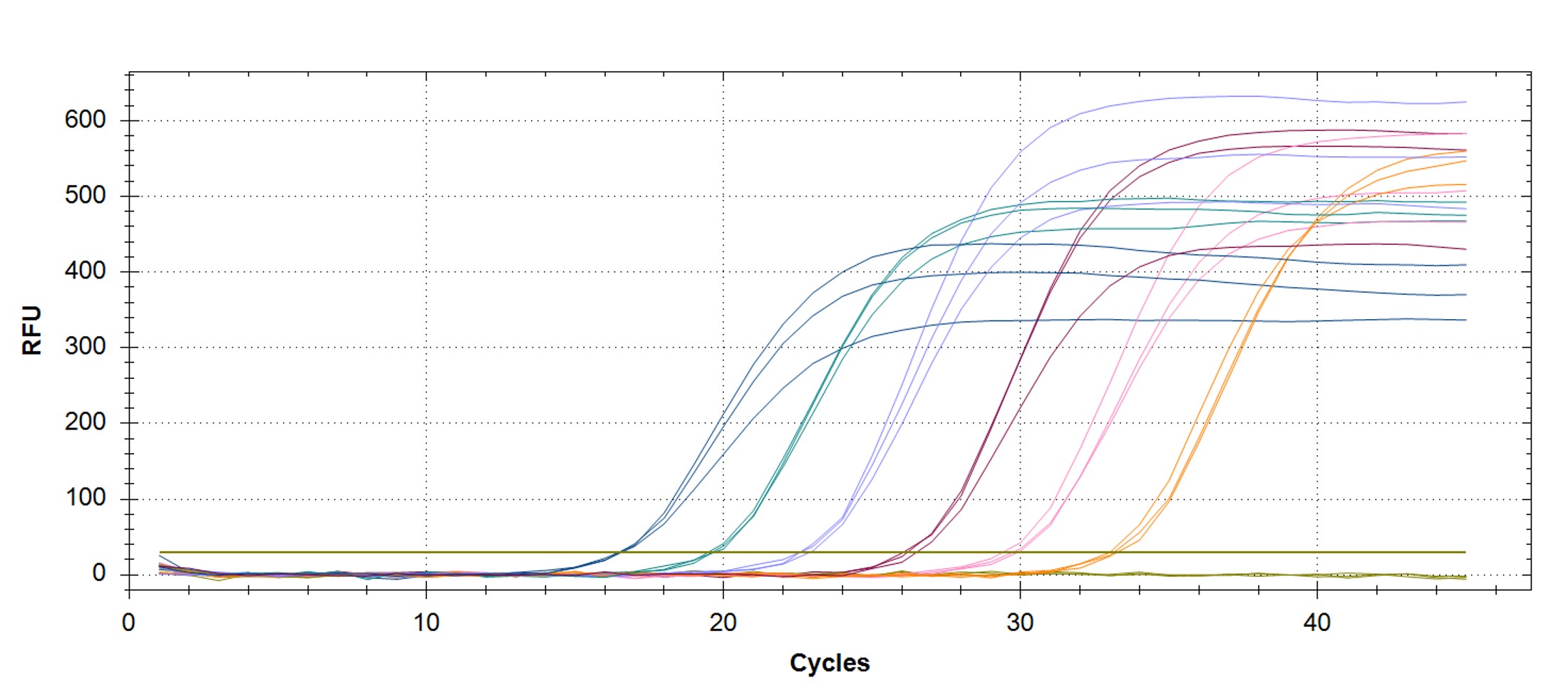
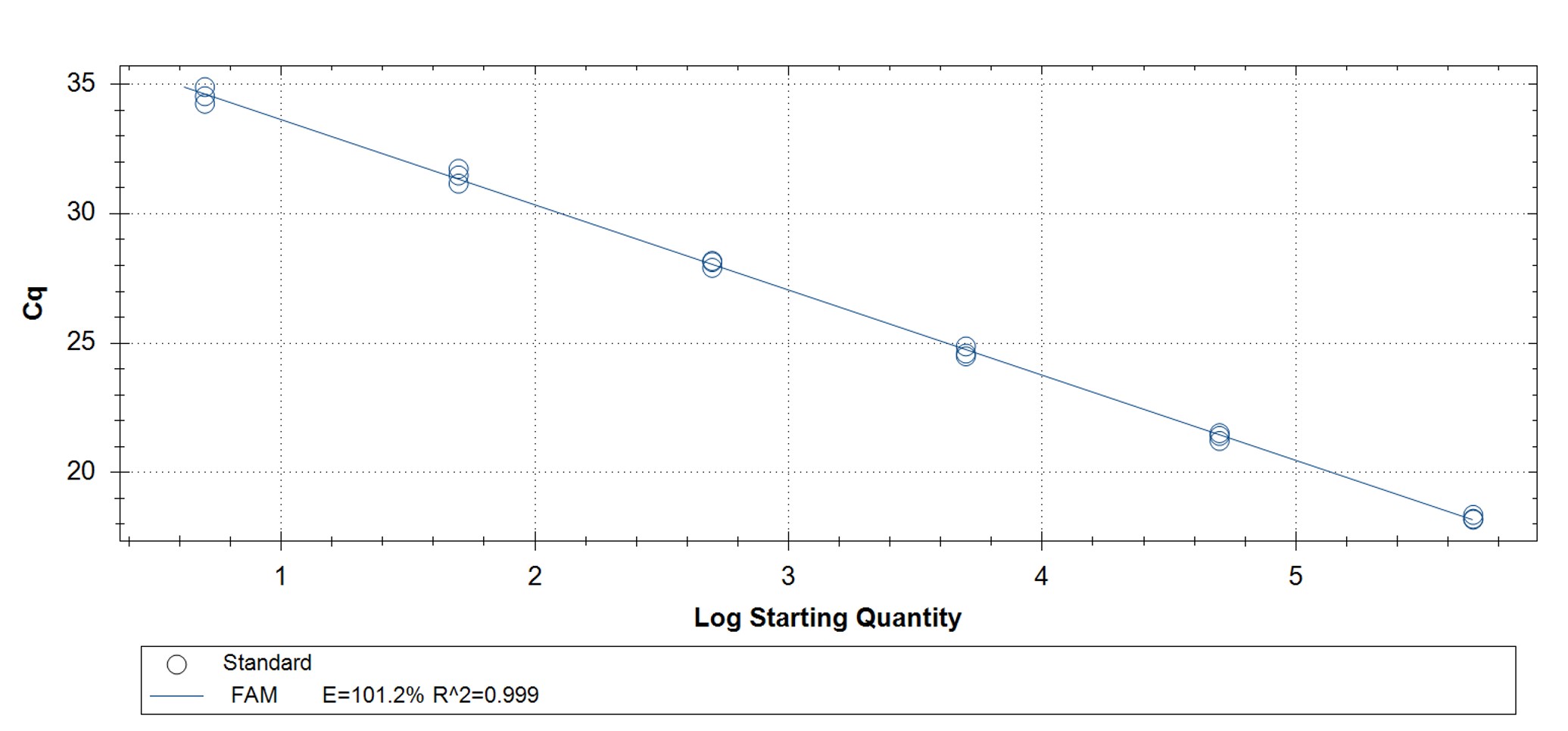
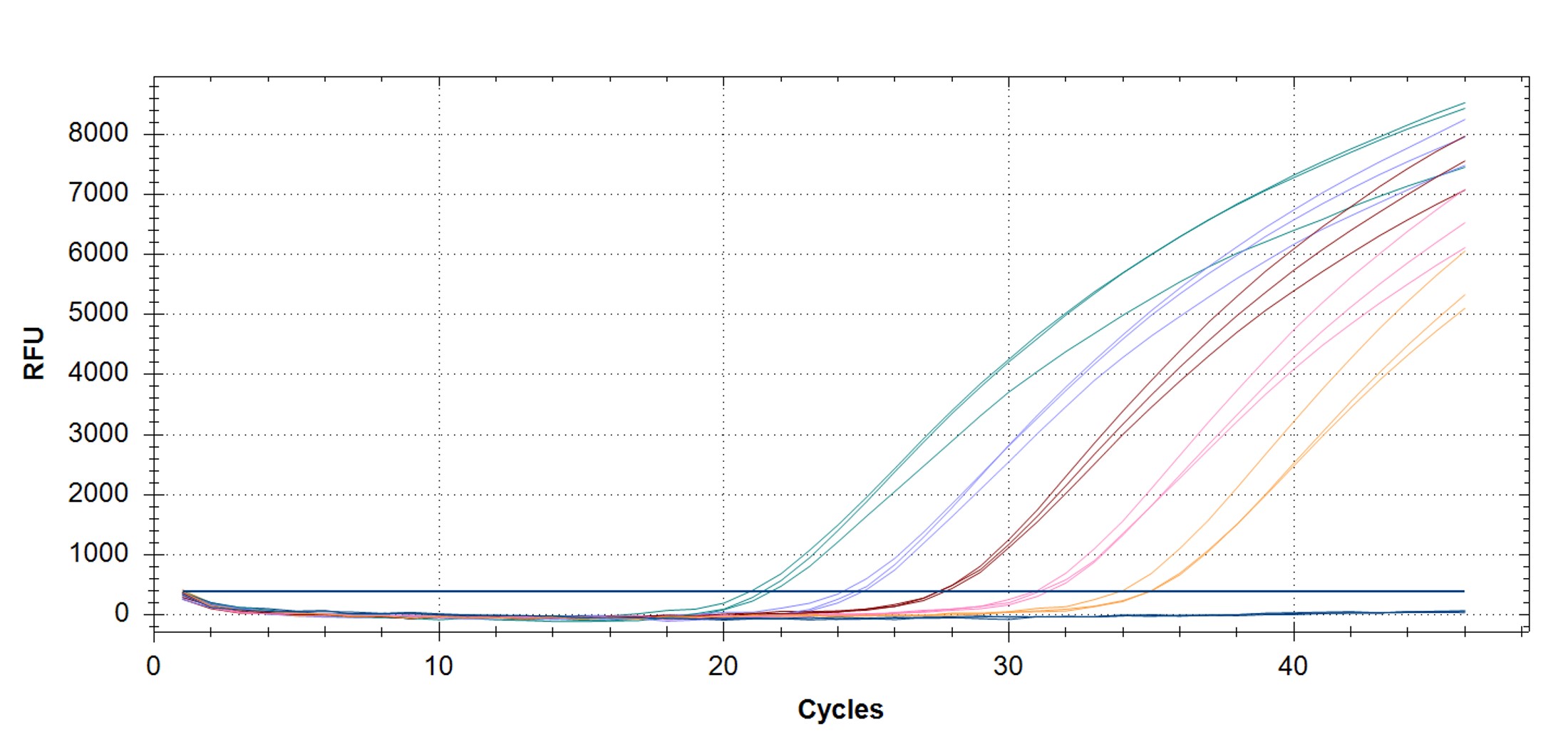
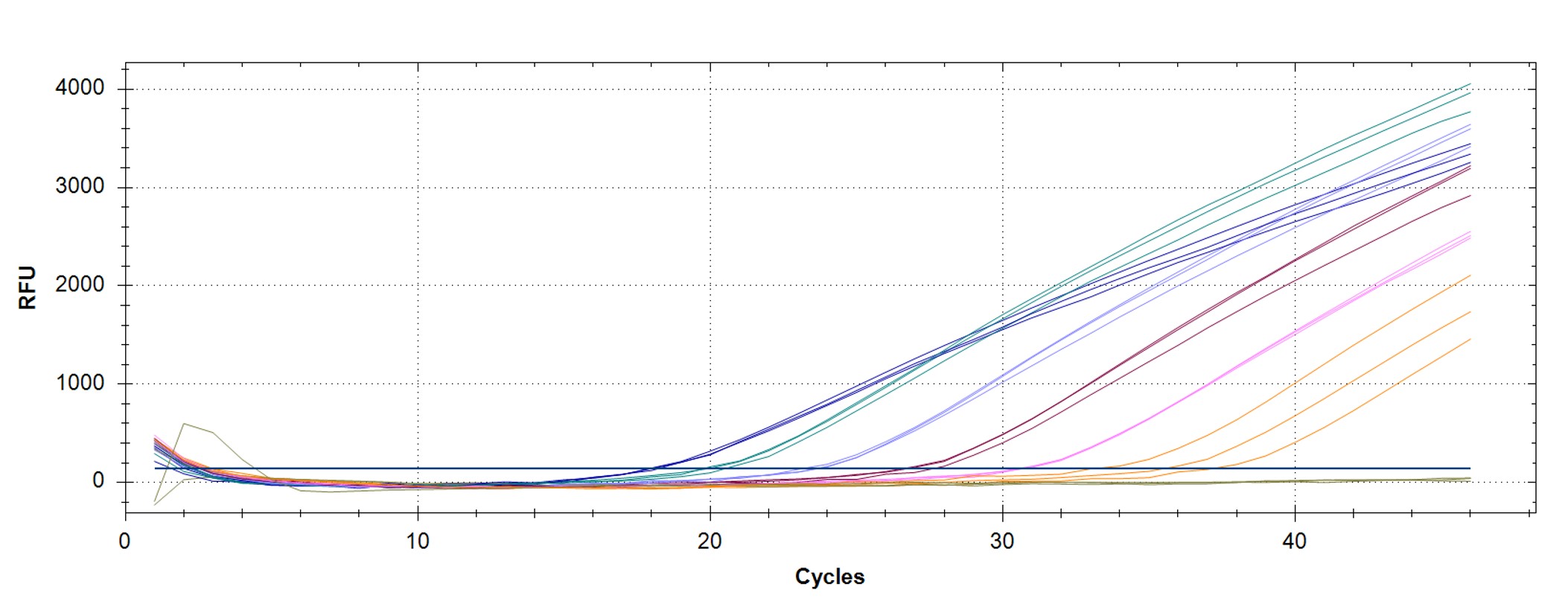
Figure 1. qPCR assay was used to verify the functionality of the synthetic molecular standard. (A) An amplification plot and (B) standard curve were generated with the hMPXV standard. The qPCR assay was performed as previously described by Maksyutov et al.1 Cycling conditions were 50°C for 2 min and 95°C for 2 min, followed by 45 cycles of 95°C for 15 sec and 63°C for 1 min. Standard curves were generated by using serial 10-fold dilutions that ranged from 5 copies/µL to 5×105 copies/µL. The synthetic DNA standard was tested in triplicate.
| A | B |
 |
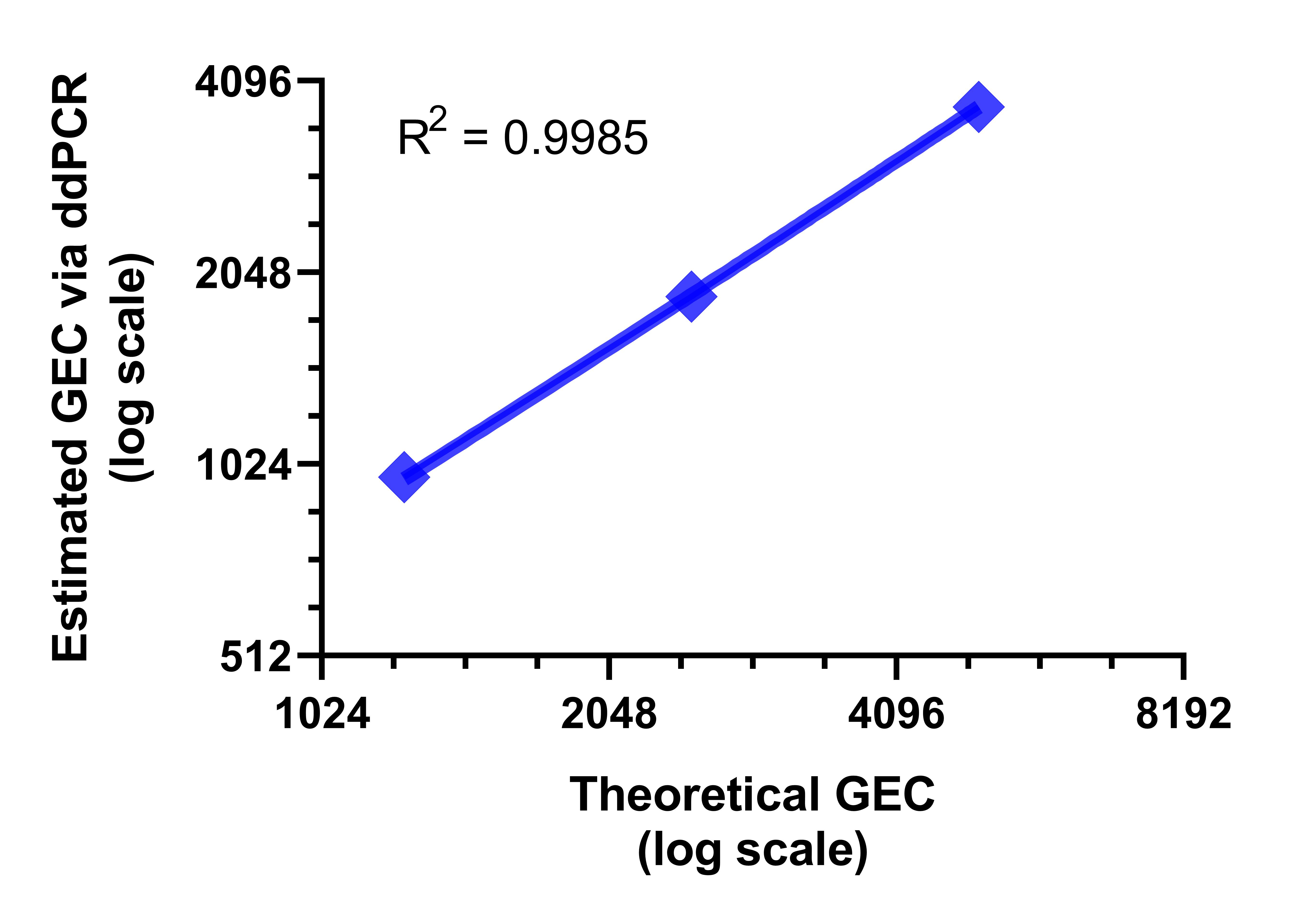 |
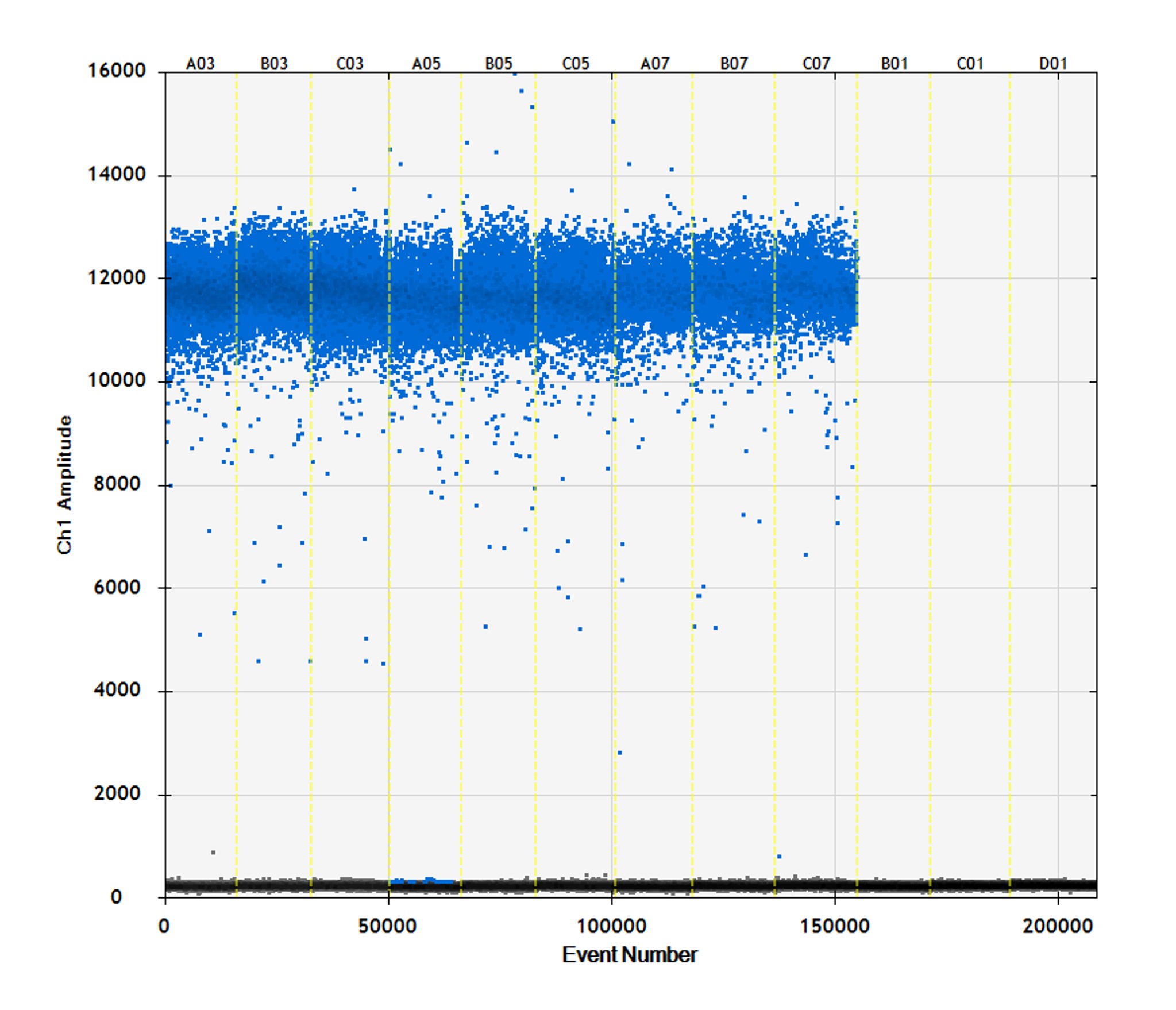
Figure 2. Absolute quantification of hMPXV via droplet digital PCR. (A) One-dimensional (1D) amplitude scatter plot of positive and negative ddPCR for three dilutions (dilution factors 100, 200, and 400) in triplicate were quantified by ddPCR by using a modified protocol of the published qPCR assay.1 ddPCR was performed as follows: initial denaturation at 95°C for 10 min, amplified 40× at 94°C for 30 sec and 60°C for 1 min, and enzyme deactivation at 98°C for 10 min. Droplets were analyzed in the QX200 droplet reader. Data were analyzed with QuantaSoft software (Bio-Rad). (B) Average calculated copy numbers/µL per dilution of stock material.
| A | B |
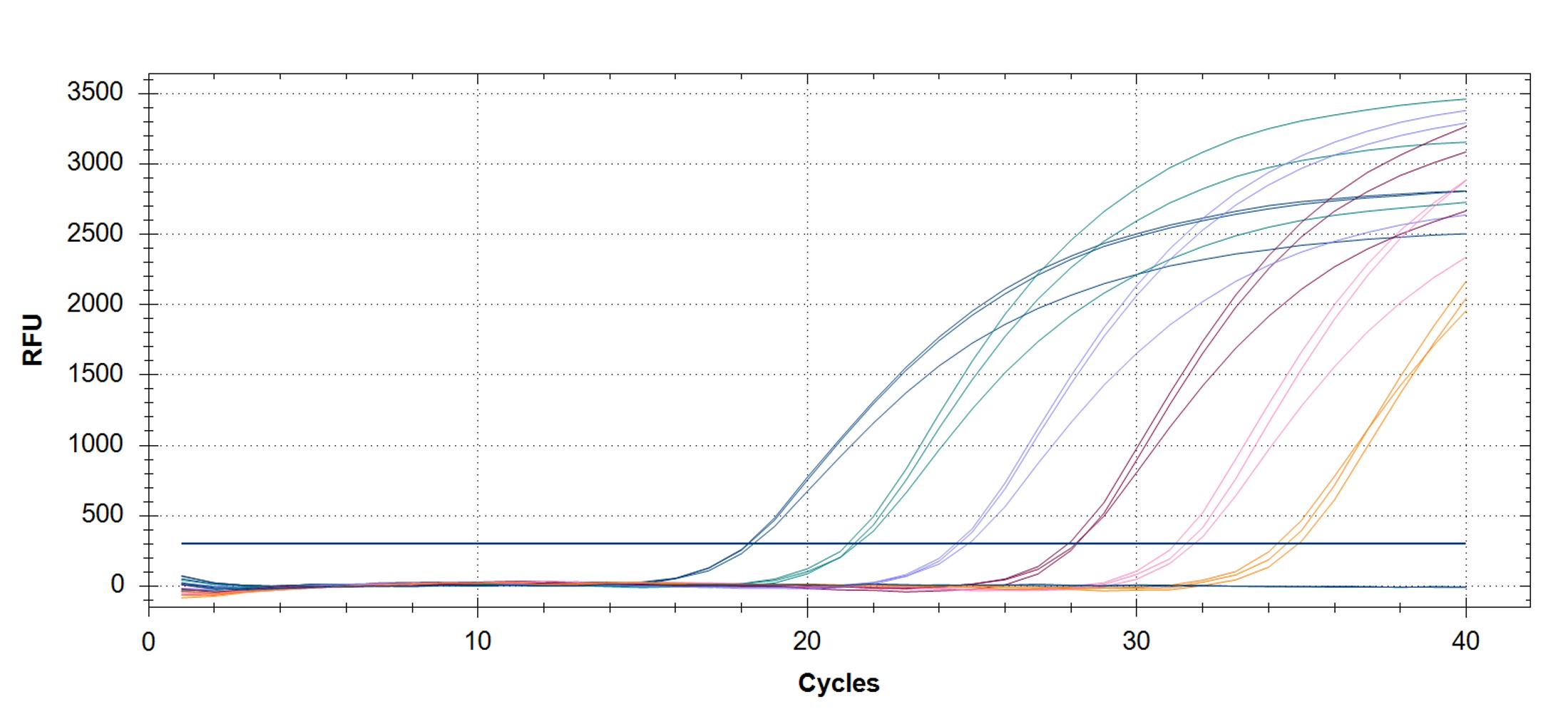 |
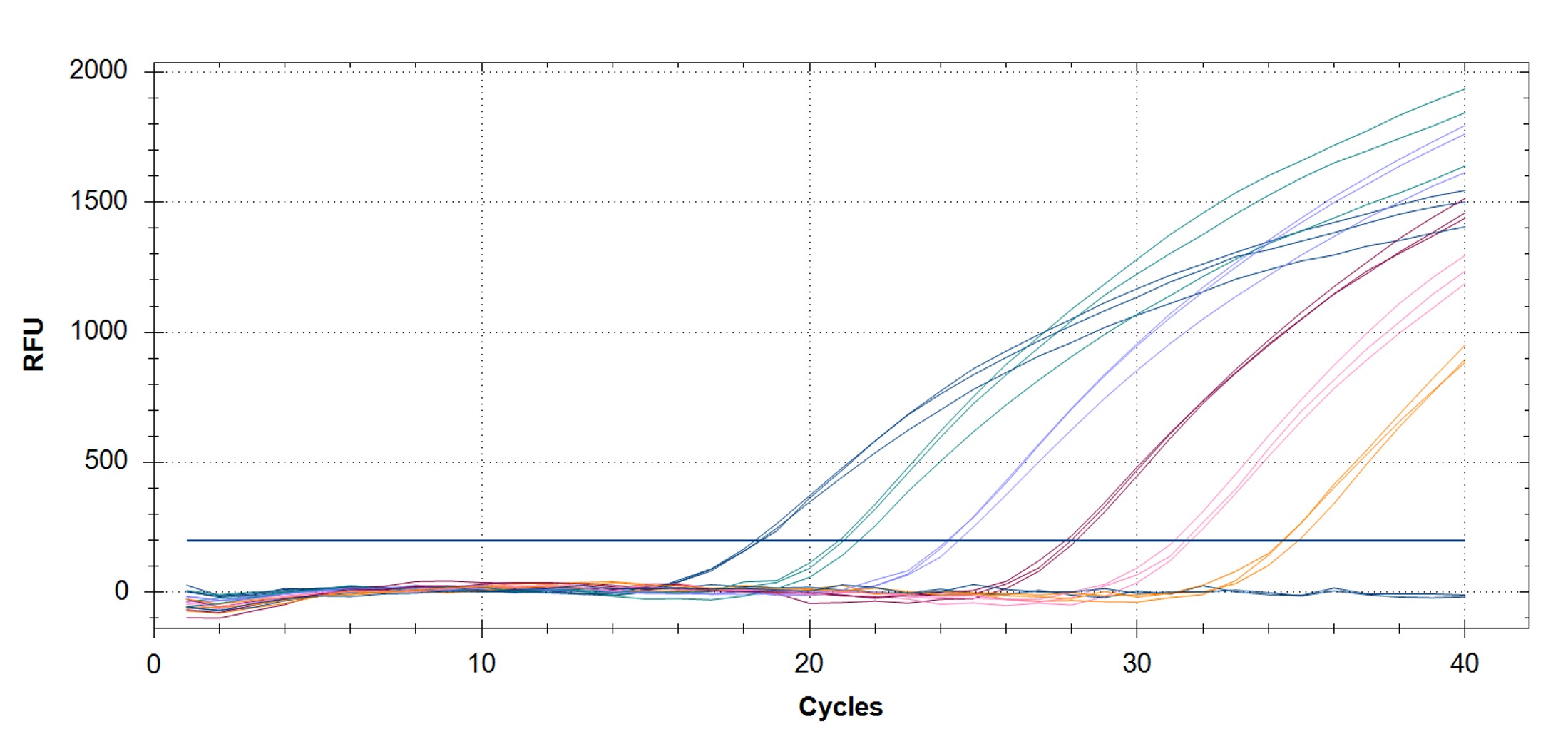
|
| C | D |
 |
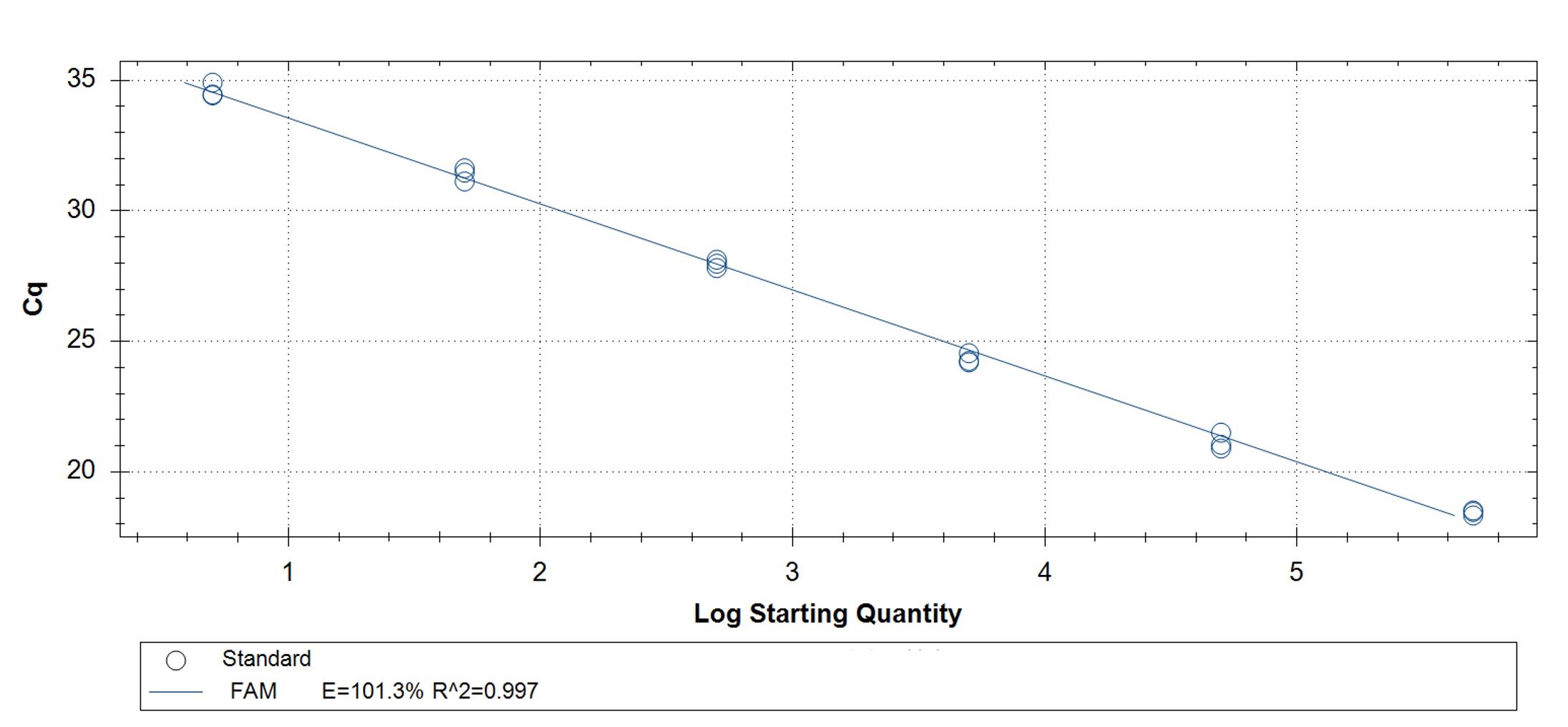 |
Figure 3. qPCR assays. (A, C) Amplification plots and (B, D) standard curves were generated with the hMPXV standard using the primers and probe from the CDC non-variola orthopoxvirus (C, D) and Monkeypox virus (A, B) generic assays. The qPCR assay was performed as previously described by CDC.2,3 Cycling conditions for the orthopox assay were 50°C for 2 min and 95°C for 20 sec, followed by 40 cycles of 95°C for 3 sec and 63°C for 30 sec. Cycling conditions for the Monkeypox virus assay were 50°C for 2 min and 95°C for 20 sec, followed by 40 cycles of 95°C for 3 sec and 60°C for 30 sec. Standard curves were generated by using serial 10-fold dilutions that ranged from 5 copies/µL to 5×105 copies/µL. The synthetic DNA standard was tested in triplicate.
| A | B |
 |
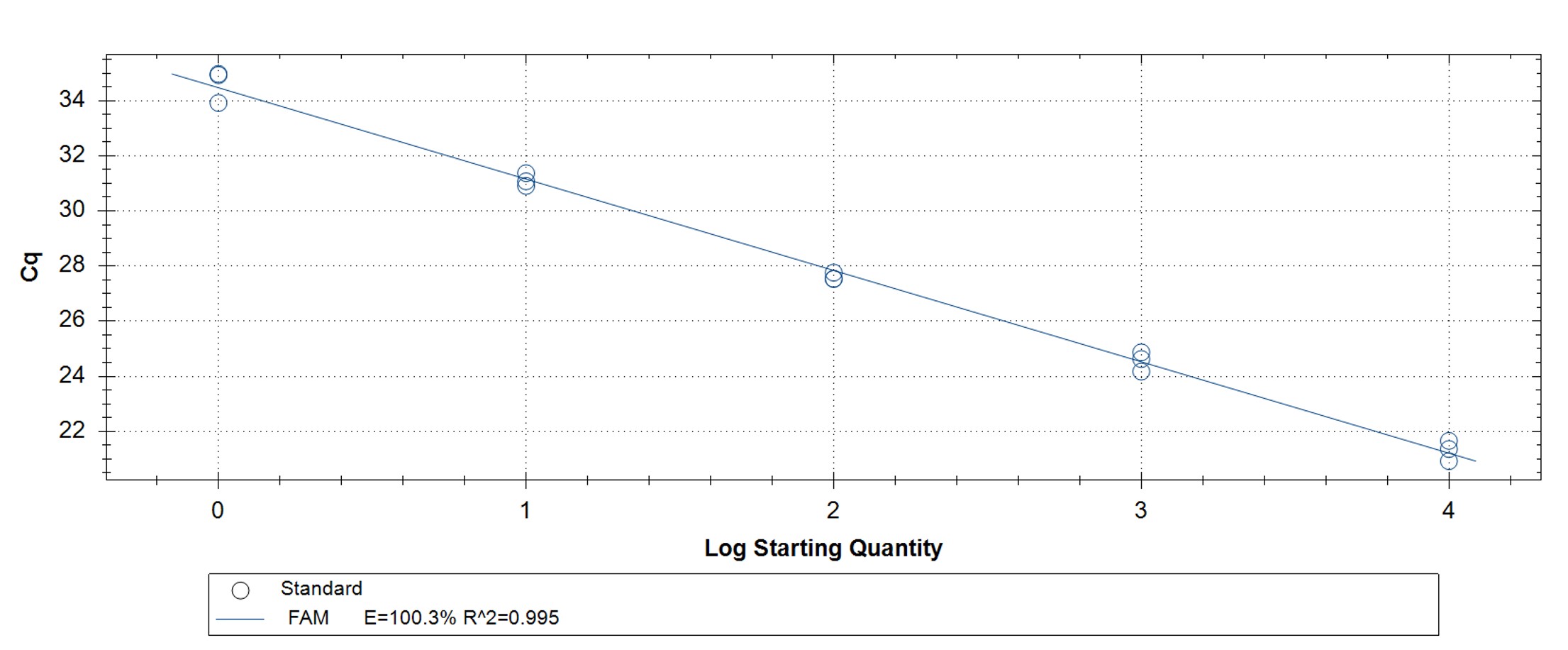 |
Figure 4. hMPXV Clade II (formerly West African clade)-specific qPCR assay to verify the functionality of the synthetic molecular standard. (A) An amplification plot and (B) standard curve were generated with the hMPXV standard. The qPCR assay was performed as previously described by Li et al.4 Cycling conditions were 95°C for 6 min, followed by 45 cycles of 95°C for 5 sec and 62°C for 20 sec. Standard curves were generated by using serial 10-fold dilutions that ranged from 5 copies/µL to 5×104 copies/µL. The synthetic DNA standard was tested in triplicate.
| A | B |
 |
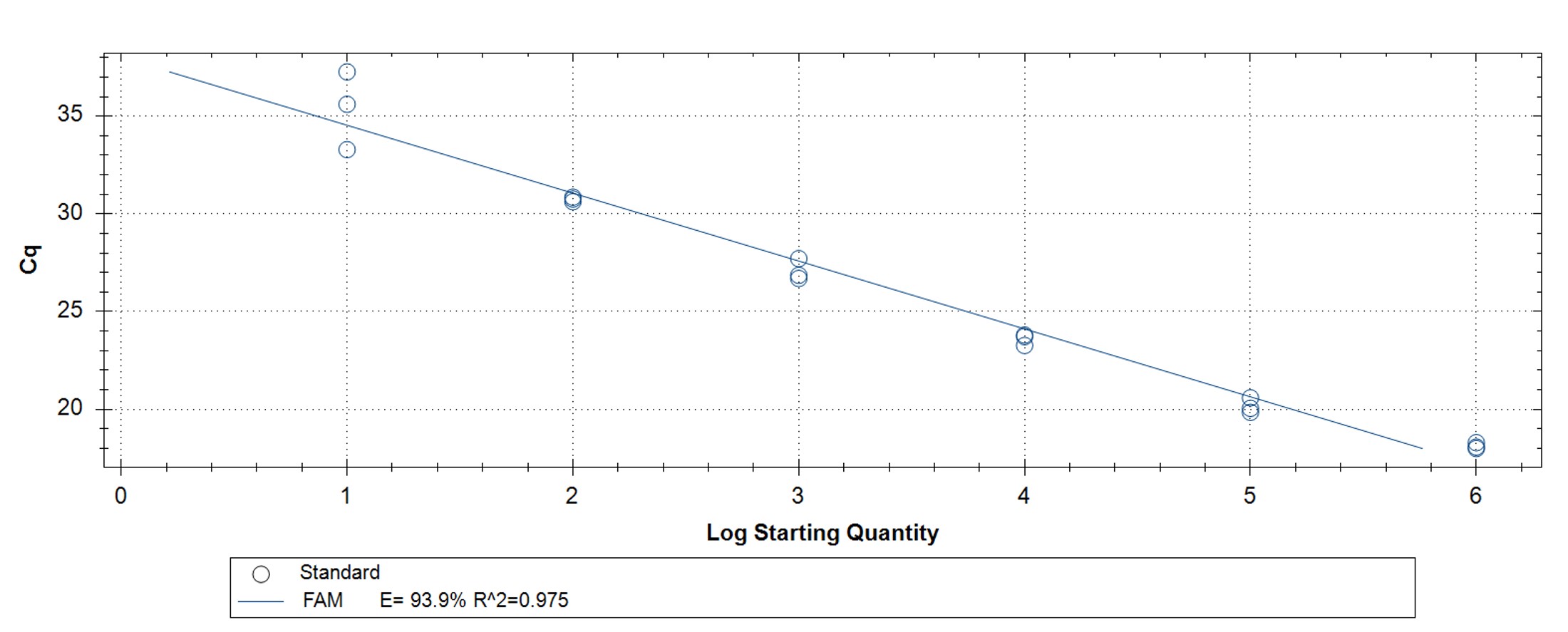 |
Figure 5. hMPXV Clade I (formerly Congo Basin/Central African clade)-specific qPCR assay to verify the functionality of the synthetic molecular standard. (A) An amplification plot and (B) standard curve were generated with the hMPXV standard. The qPCR assay was performed as previously described by Li et al.4 Cycling conditions were 95°C for 6 min, followed by 45 cycles of 95°C for 5 sec, 60°C for 10 sec, and 72°C for 20 sec. Standard curves were generated by using serial 10-fold dilutions that ranged from 5 copies/µL to 5×105 copies/µL. The synthetic DNA standard was tested in triplicate.
Conclusion
Our proof-of-concept data demonstrates that the ATCC quantitative synthetic molecular standard for hMPXV can be used as a control for assay development, verification, and validation. The standard was manufactured under ISO 13485 guidance and can be used to determine the microbial load of unknown mpox samples through the generation of a standard curve. The standard is compatible with numerous published assays1,2,3,4 and exhibited minimal variability, as evident from the slope and R2 values. Assays performed well with as low as 5 GEC per reaction. Taken together, this standard provides well-characterized controls for viral detection and quantification.
Like all ATCC synthetic molecular standards, ATCC VR-3270SD is a quantitative and stabilized material that can be safely handled under BSL-1 conditions. This standard can be used in a variety of applications, including the generation of a standard curve for qPCR, as a positive control for RT-PCR assays, assay verification and validation studies, monitoring assay-to-assay and lot-to-lot variation, and molecular diagnostics assay development.
Download a PDF of this application note
Download now
References
- Maksyutov RA, Gavrilova EV, and Shchelkunov SN. Species-specific differentiation of variola, monkeypox, and varicella-zoster viruses by multiplex real-time PCR assay. J Virol Methods 236: 215-220, 2016. PubMed: 27477914
- Centers for Disease Control & Prevention Poxvirus & Rabies Branch (PRB). Test Procedure: Non-variola Orthopoxvirus Generic Real-Time PCR Test. Rev. No. 02, 2022.
- Centers for Disease Control & Prevention Poxvirus & Rabies Branch (PRB). Test Procedure: Monkeypox virus Generic Real-Time PCR Test. Rev. No. 01, 2022.
- Li Y, Zhao H, Wilkins K, Hughes C, Damon IK. Real-time PCR assays for the specific detection of monkeypox virus West African and Congo Basin strain DNA. J Virol Methods 169: 223–227, 2010. PubMed: 20643162
