Shamaila Ashraf, PhD; Prasanthi Bandi, MS; Brian Chase, MS; and Dev Mittar, PhD
ATCC, Manassas, VA
Introduction
Development of the synthetic molecular standards
Generation of standard curves from second-generation NoV synthetic molecular standards
Comparison of first-generation and second-generation synthetic molecular standards
Quantification of the NIBSC working reagents for NoV-GI and NoV-GII using ATCC second-generation synthetic molecular standards
Conclusion
References
Abstract
ATCC has improved existing first-generation qualitative synthetic molecular standards for Norovirus genogroups I and II (NoV-GI and NoV-GII, respectively) through the development of 2 new quantitated synthetic molecular standards. These second-generation synthetic RNA standards are synthesized under ISO 13485 guidance and comprise short conserved fragments representing the NoV genome. Further, they are compatible with numerous published quantitative reverse transcription polymerase chain reaction (qRT-PCR) assays, can be handled at biosafety level one (BSL-1), are stable, and can be shipped internationally.
Download a PDF of this application note
Download NowIntroduction
NoVs are the most common cause of epidemic gastroenteritis, accounting for 95% of viral gastroenteritis outbreaks worldwide. These viruses are difficult to detect due to their genetic heterogeneity and unculturable nature. Currently, the principle detection method utilized by diagnostic laboratories is qRT-PCR. The accuracy of a qRT-PCR assay relies on the generation of a standard curve using a positive control with a known genome copy number. In addition, clinical laboratories require an independent control to monitor the accuracy of their assays. Since Norovirus cannot be grown in cell culture, the acquisition of such positive control material proves challenging. To circumvent these issues, ATCC has developed quantitative synthetic molecular standards that include conserved sequences from NoV-GI and NoV-GII for the detection and quantification of NoV from either clinical, food, or environmental samples (Table 1).
These novel synthetic molecular standards are an improvement over existing first-generation qualitative synthetic molecular standards for NoV-GI and NoV-GII in that they are synthesized under ISO 13485 guidance, quantified by Droplet Digital PCR (ddPCR, Bio-Rad), and stabilized using a proprietary stabilization matrix. Further, the improved design of these second-generation constructs enables compatibility with additional molecular assays for the detection and quantification of NoV. In this study, we used the second-generation synthetic molecular standards for NoV-GI and NoV-GII to generate standard curves using 2 different primer sets: one used by the laboratories in the CaliciNet1 and another recommended by the European Committee for Standards (CEN/TC/WG6/TAG4) working group.2 Furthermore, we used the respective standard curves to quantify the working reagents for NoV-GI and NoV-GII from National Institute for Biological Standards and Control (NIBSC) using the above primer sets.
Table 1. ATCC synthetic NoV molecular standards
| ATCC number | Designation |
|---|---|
| VR-3234SD | Synthetic NoV-GI RNA |
| VR-3235SD | Synthetic NoV-GII RNA |
Development of the synthetic molecular standards
The quantitative synthetic molecular standards for NoV were manufactured under ISO 13485 guidance and include respective fragments from the ORF1 and ORF2 junction for NoV-GI and NoV-GII, and an additional fragment from the VP2 region for NoV-GII. Both synthetic RNA standards were quantified by ddPCR in order to package precise copies of RNA. Given the inherent labile nature of RNA, a stabilization matrix was added to the quantitated RNA preparations.
Generation of standard curves from second-generation NoV synthetic molecular standards
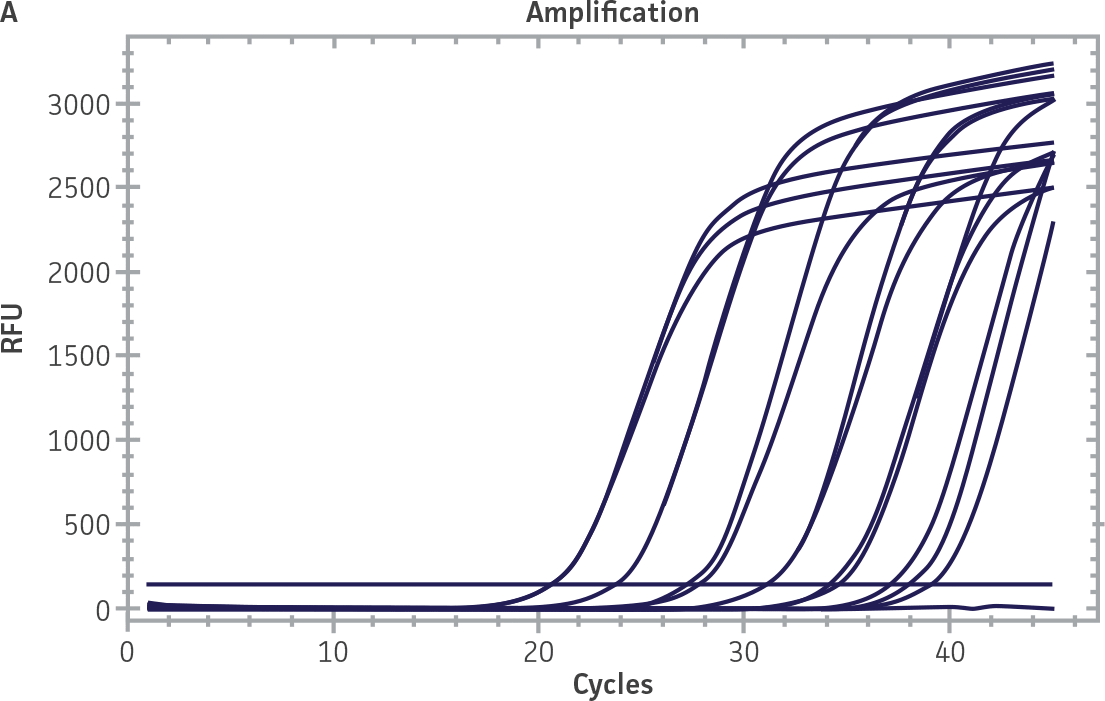
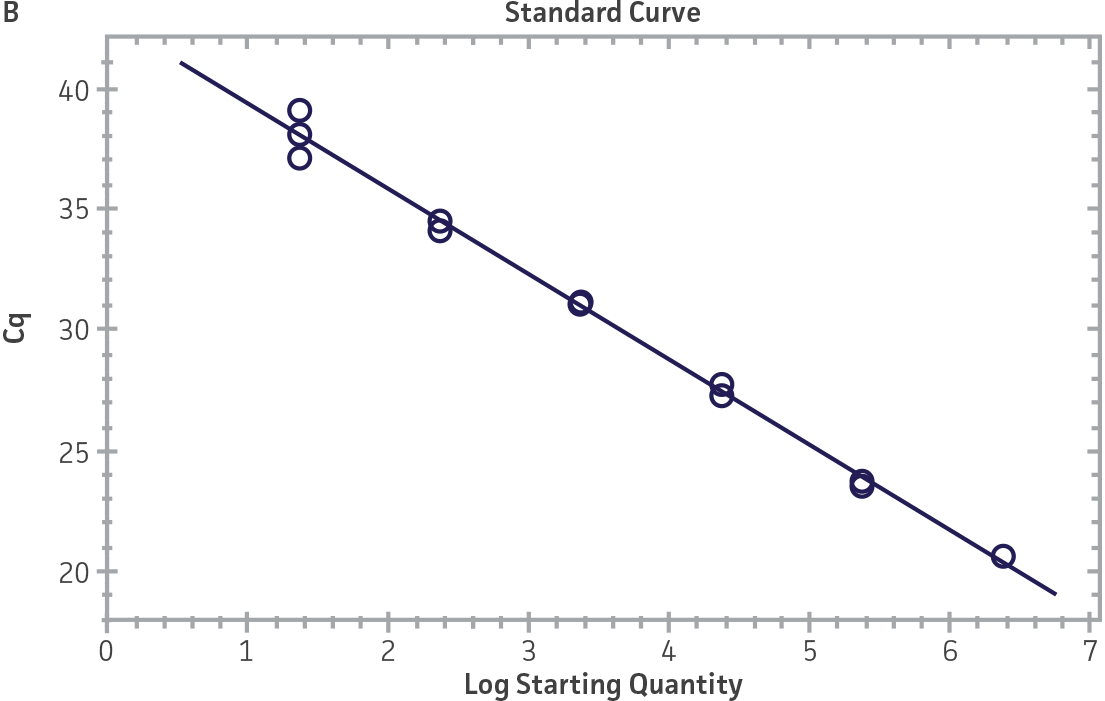
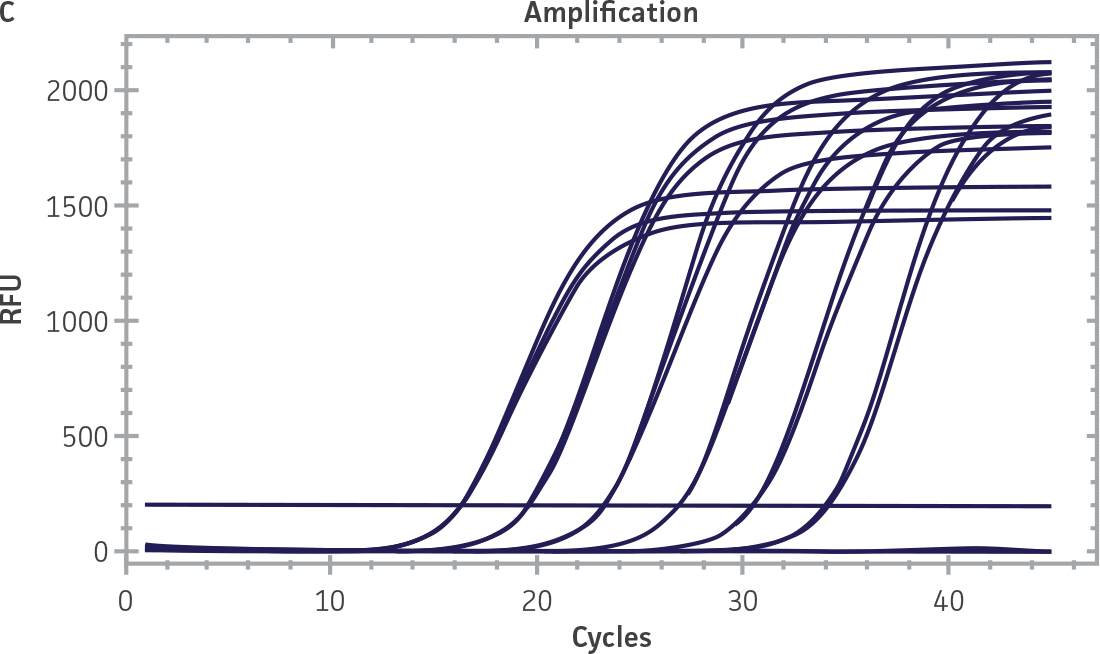
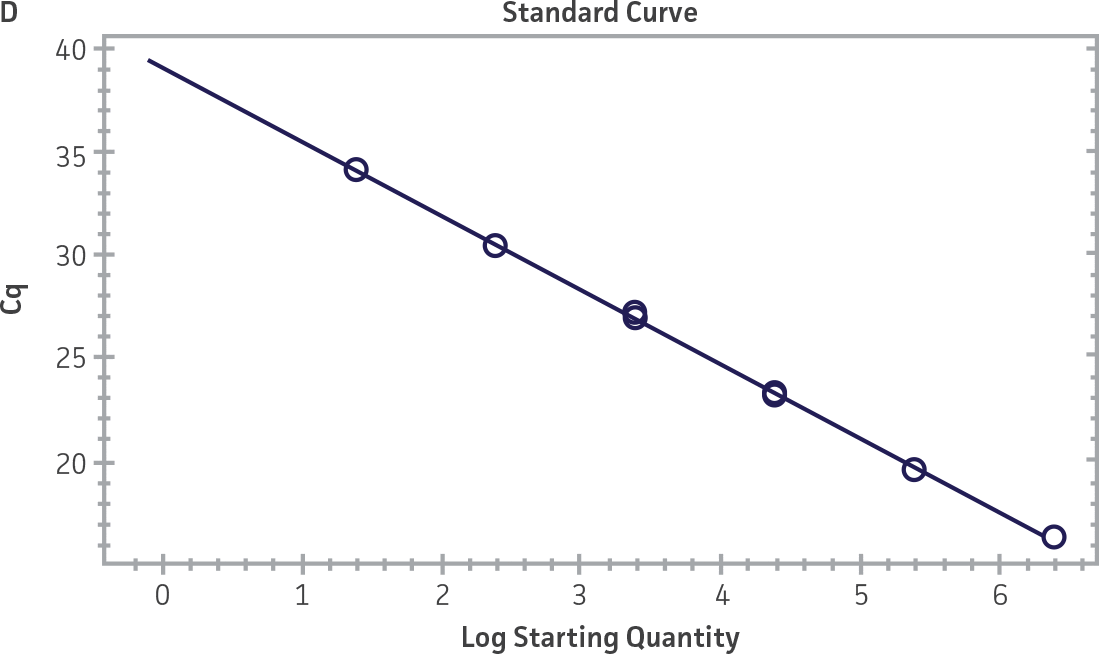
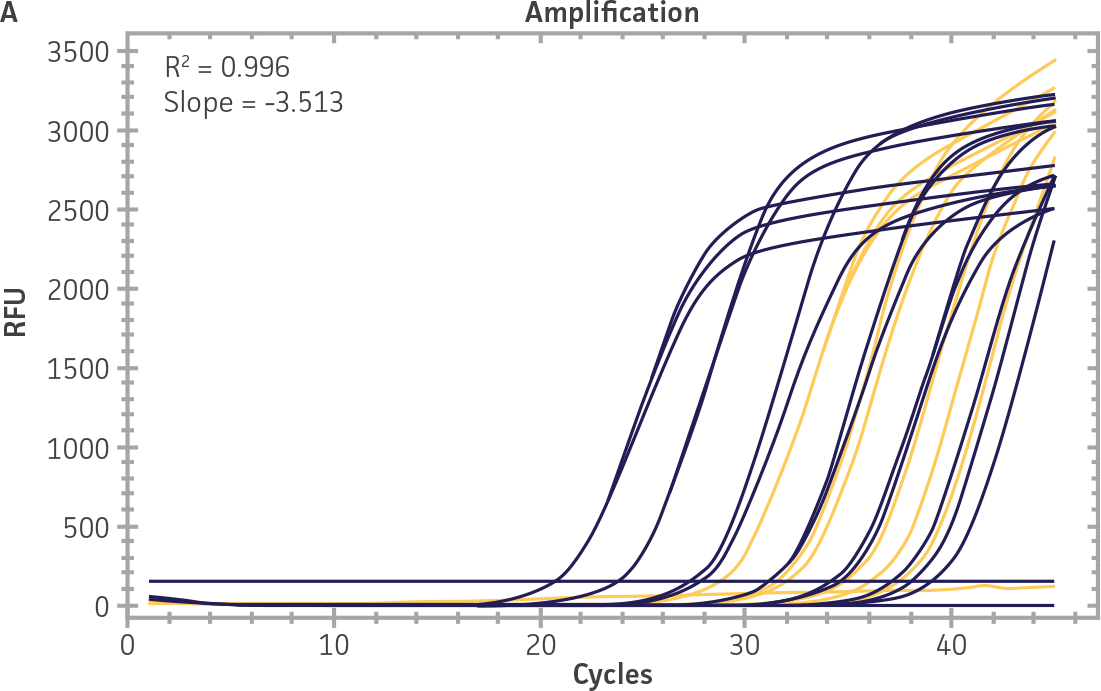
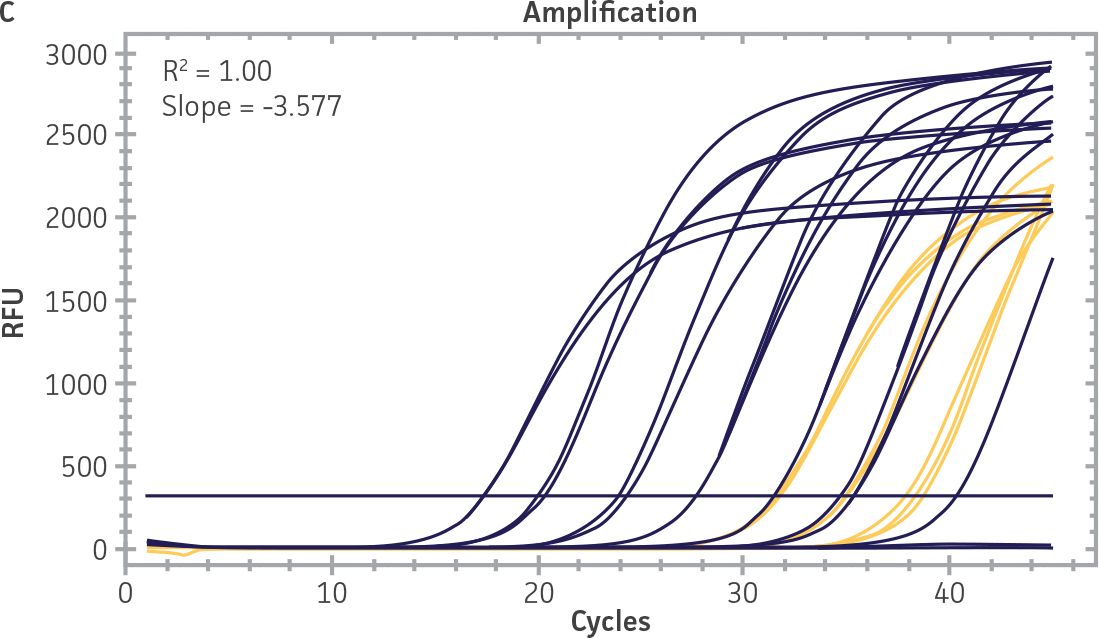
Standard curves were generated using serial tenfold dilutions of the synthetic RNA standards, ranging from 20 copies to 2 × 106 copies/reaction (Figure 1). Standards were tested in triplicate wells using the CFX96 Real-Time PCR Detection System (Bio-Rad). The primer and probe sets from the Real-Time RT-PCR assay used by laboratories in CaliciNet1 and an assay recommended CEN/TC/WG6/TAG4 group2 were employed to examine each individual NoV genotype using the following conditions. Cycling conditions for all primer sets were 50°C for 15 min and 95°C for 2 min, followed by 45 cycles of 95°C for 15 sec and 60°C for 60 sec for both the NoV-GI and NoV-GII assays. RNA samples and standards were tested in triplicate. The relative fluorescence unit (RFU) baseline threshold was set and genome copy numbers calculated using CFX Manager 3.0 Software (Bio-Rad). From this analysis, it was determined that the second-generation synthetic NoV molecular standards are compatible with both primer sets over a broad linear dynamic range and exhibit minimal variability as evident from slope and R2 values (Figure 1E).
 |
 |
 |
 |
E
| Primer and probe | NoV-GI | NoV-GII | |
|---|---|---|---|
| CaliciNet Assay1 | Slope R2 |
-3.513 0.996 |
-3.577 1.00 |
| CEN/TC/WG6/TAG4 Assay2 | Slope R2 |
-3.692 0.987 |
-3.625 0.998 |
Figure 1. Generation of Standard Curves from Second-Generation NoV Synthetic Molecular Standards. (A&C) Amplification plots and (B&D) standard curves generated using the second-generation NoV-GI and NoV-GII synthetic molecular standards (ATCC VR-3234SD and ATCC VR-3235SD, respectively) with the CaliciNet primer and probe set.1 (E) The slope and R2 values generated using the second-generation NoV-GI and NoV-GII synthetic molecular standards with the primer and probe sets from the CaliciNet1 and CEN/ TC/WG6/TAG42 real-time RT-PCR assays.
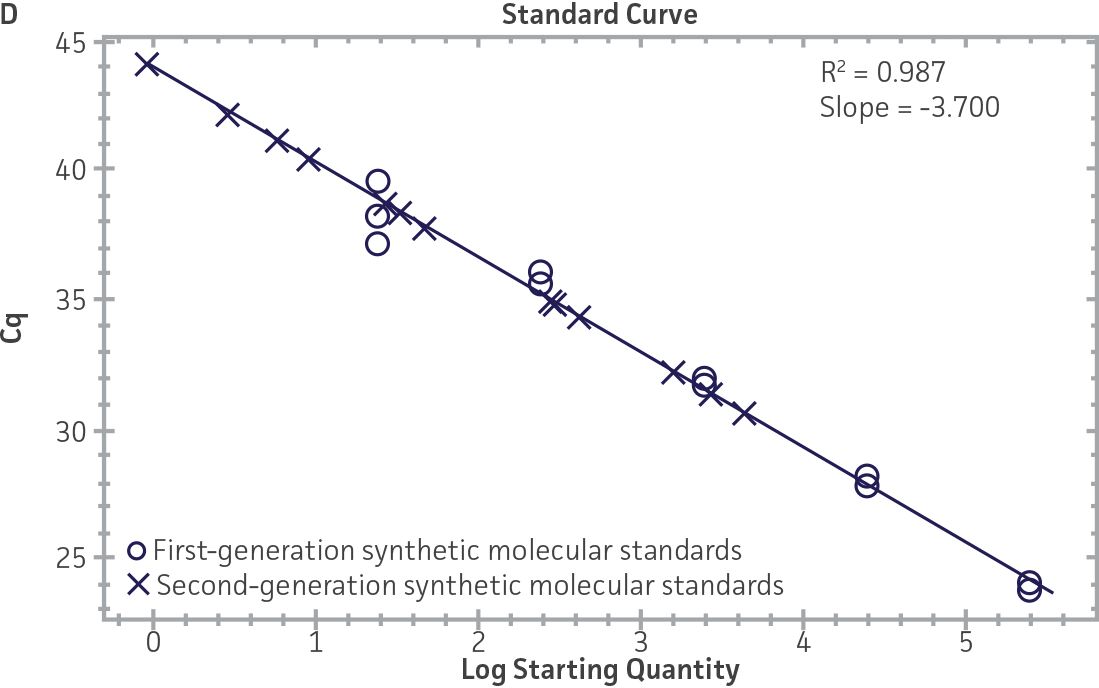
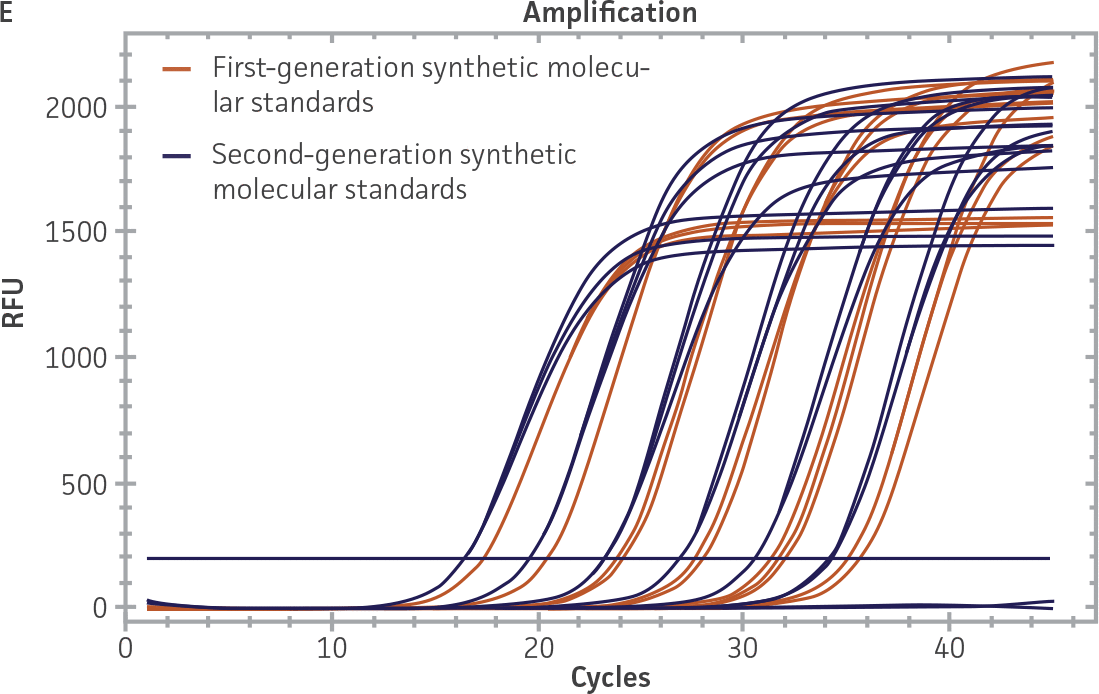
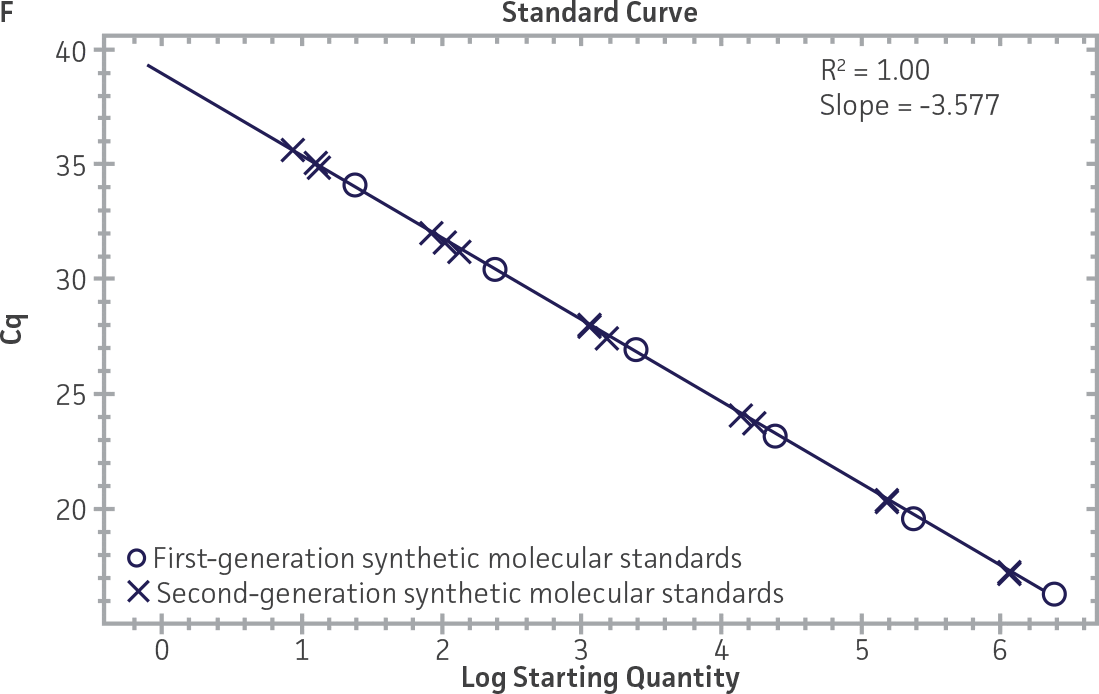
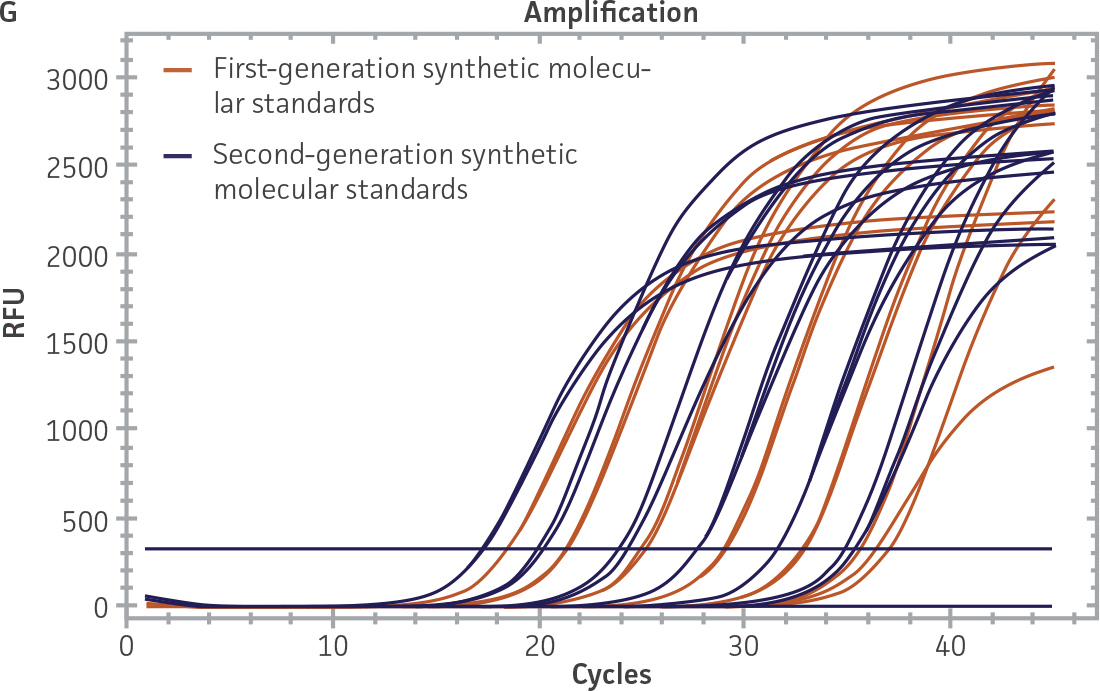
Comparison of first-generation and second-generation synthetic molecular standards
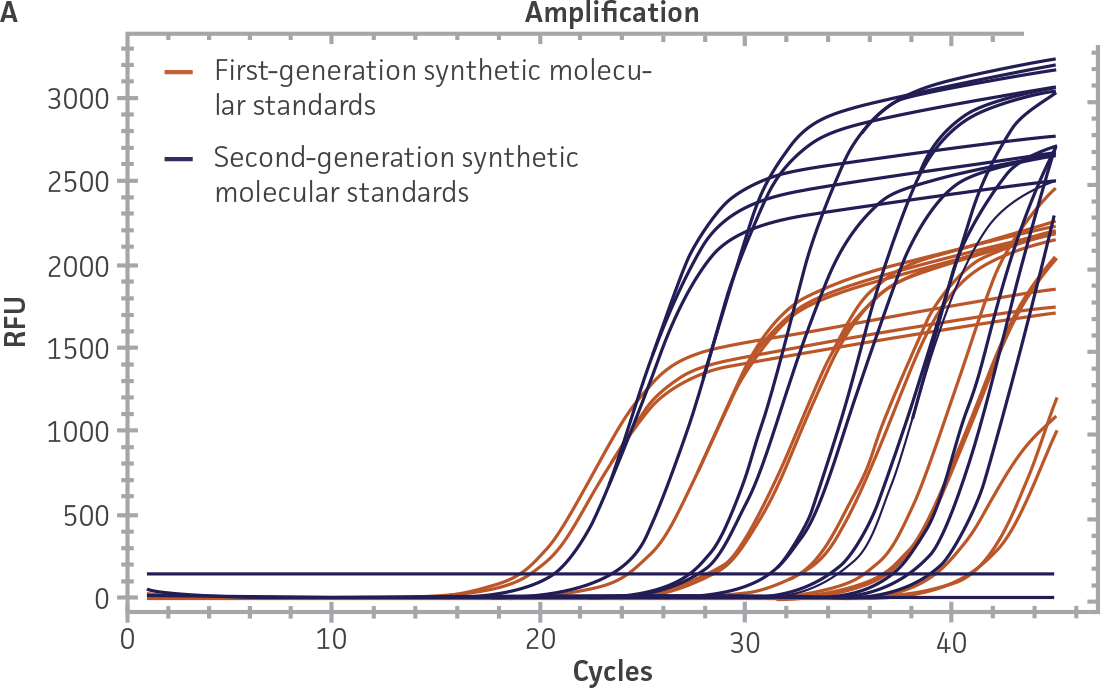
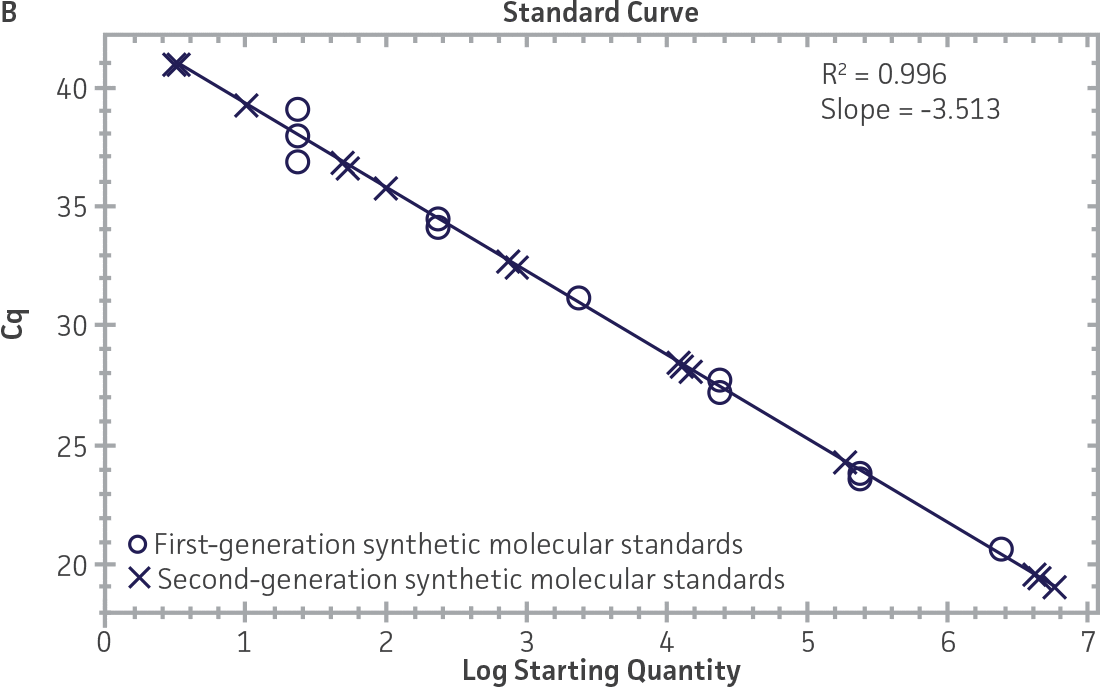
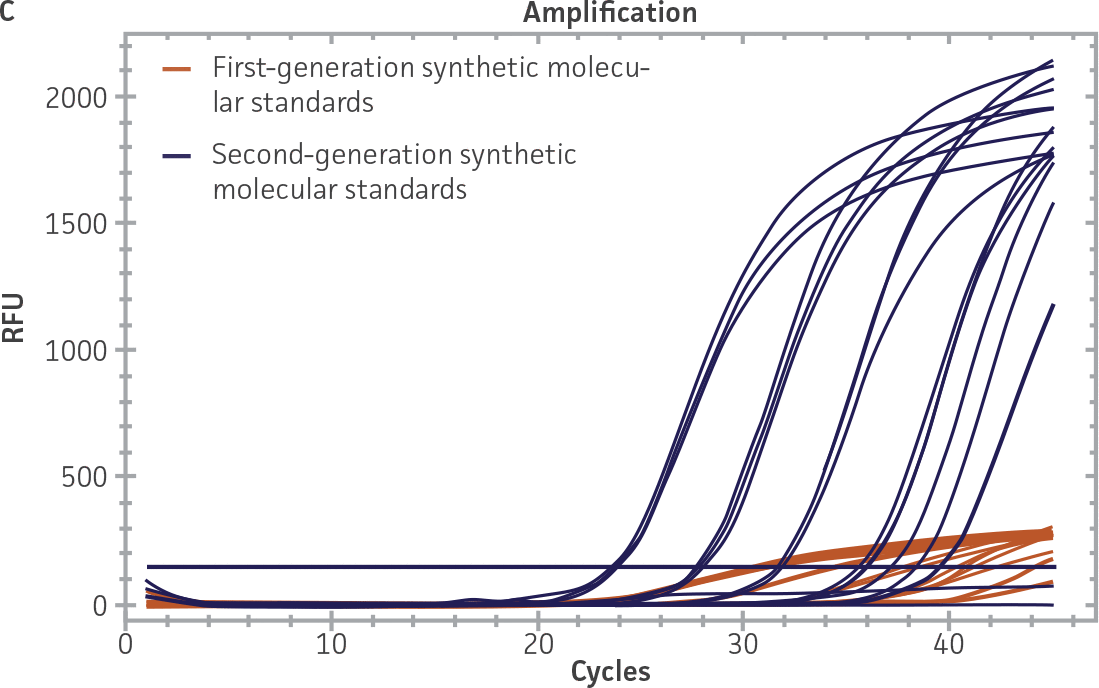
First-generation and second-generation synthetic molecular standards were compared using the primer and probe sets from both the CaliciNet1 and CEN/TC/WG6/TAG42 assays (Figure 2). Standard curves were generated using serial tenfold dilutions of the synthetic RNA standards and were tested in triplicate wells using the CFX96 Real-Time PCR Detection System as described above. From this analysis, it was determined that the first-generation molecular standards were not compatible with the primer and probe set used in the CEN/TC/WG6/TAG4 assay2 for NoV-GI detection (Figure 2C), whereas the improvement in design of the newer second-generation constructs enabled consonance with both assays.
 |
 |
 |
 |
 |
 |
 |
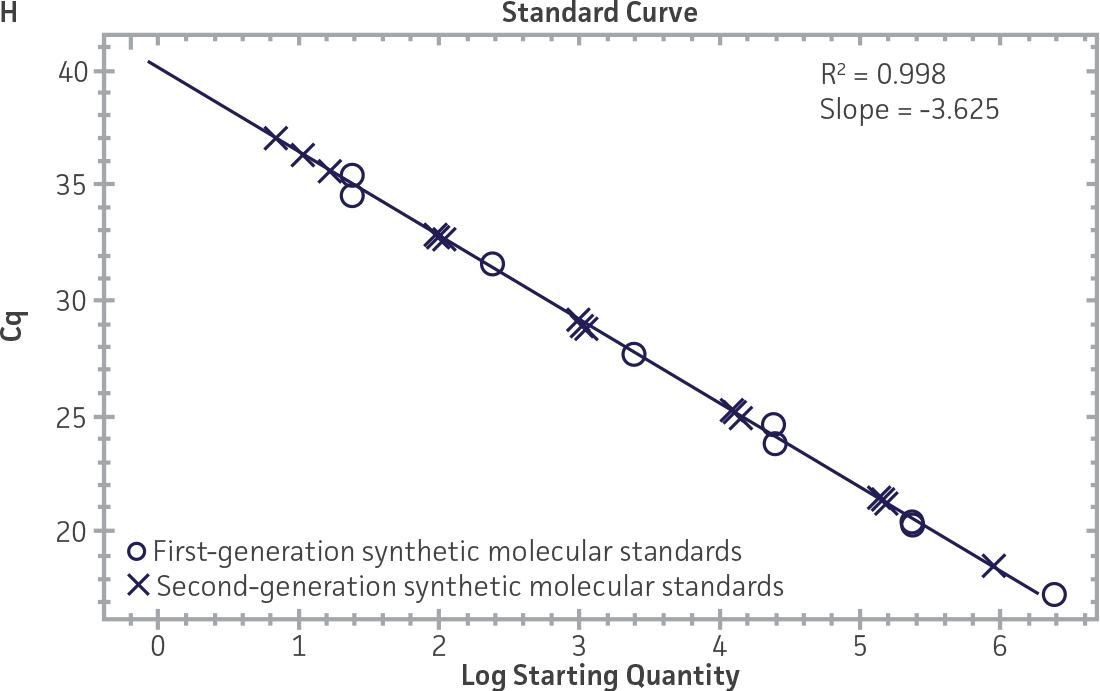 |
Figure 2. Comparison of First-Generation and Second-Generation Synthetic Molecular Standard. (A&C) Amplification plots and B&D) standard curves generated using the first- and second-generation NoV-GI molecular standard ATCC VR-3199SD and ATCC VR-3234SD, respectively, versus the (E&G) amplification plots and (F&H) standard curves generated using the first- and second-generation NoV-GII synthetic molecular standards ATCC VR-3200SD and ATCC VR-3235SD, respectively. Amplification plots (A&E) and standard curves (B&F) were generated using the primer and probe set used in the CaliciNet assay.1 Amplification plots (C&G) and standard curves (D&H) were generated using the primer and probe set used in the CEN/TC/WG6/TAG4 assay.2
Quantification of the NIBSC working reagents for NoV-GI and NoV-GII using ATCC second-generation synthetic molecular standards
As a proof of concept, we used the respective standard curves generated using the second-generation synthetic molecular standards paired with either the CaliciNet1 or CEN/TC/WG6/TAG4 assay2 primer and probe sets to quantify the NIBSC working reagents for NoV-GI and NoV-GII (Figure 3). Viral RNA from the NoV-GI and NoV-GII working reagents was extracted using the QIAamp Viral RNA Mini Kit (QIAGEN). The NIBSC working reagents were assayed in triplicate wells as undiluted samples and diluted 1:10 and 1:100 for the qRT-PCR assay. The average from the triplicate wells from undiluted samples was used to calculate the quantities of the NoV genome as 2.3 × 103 copies /μL for NoV-GI and 46.8 copies /μL for NoV-GII.
 |
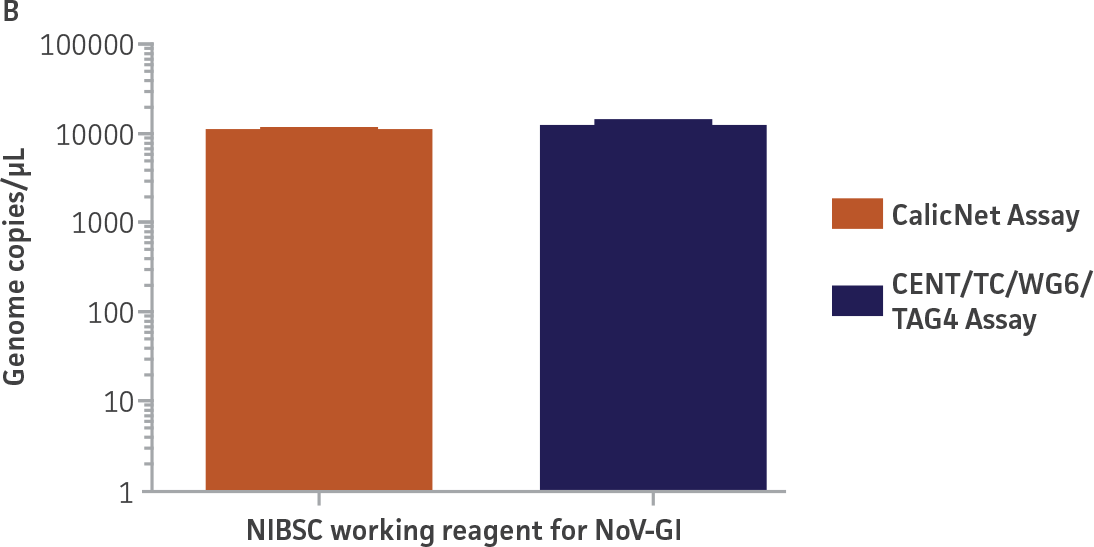 |
 |
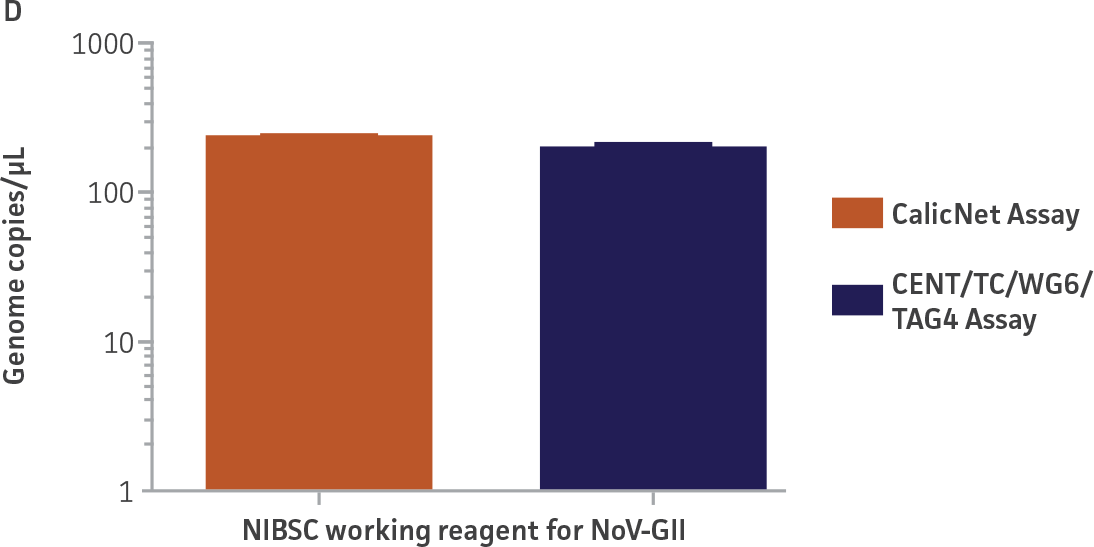 |
Figure 3. Quantification of the NIBSC Working Reagents for NoV-GI and NoV-GII Using ATCC Second-Generation Synthetic Molecular Standards. An example of a qRT-PCR amplification plot showing the second-generation (A) NoV-GI and (C) NoV-GII synthetic molecular standards (Purple) and the working reagent from NIBSC (Red). (B&D) Quantitation of the NIBSC working reagent for NoV-GI and NoV-GII samples, respectively, using the CaliciNet1 and CEN/TC/WG6/TAG4 assays.2
Conclusion
The second-generation NoV synthetic molecular standards have been significantly improved over the first-generation synthetic molecular standards. The quantitative nature of these standards directly allows for the generation of standard curves for determining NoV genome copies from unknown samples. Moreover, the improved design of the synthetic construct provides the ability to work with additional molecular assays for the detection and quantification of NoV.
Download a PDF of this application note
Download Now