Authors: Megan Bumann, BS; Katherine Burgomaster, MS; and Dev Mittar, PhD
Introduction
Reporter Signal Detection
Chromogenic Medium
Growth Effects
Plasmid Stability
Conclusion
References
Abstract
ATCC has developed green fluorescent protein (GFP)-labeled strains representing several clinically relevant Shiga toxin-producing Escherichia coli (STEC) serogroups associated with foodborne disease. These products are compatible with various fluorescence-based detection technologies such as a UV transilluminator, microscopy, and flow cytometry, enabling rapid and easy detection of strains. Here, we evaluate their use as positive controls.
Download a PDF of this application note
Download NowIntroduction
Growing concern over bacterial food contamination has led to increased examination of food testing protocols in today’s industry. Currently, the use of bacterial strains as positive controls in testing protocols is not widely practiced for fear of cross-contaminating samples. Due to ongoing scrutiny of food testing methodology and growing regulations under the Food and Drug Administration (FDA) Food Safety Modernization Act, it is imperative to have control strains with unique, easily detectable traits that distinguish positive control strains from actual food contaminants, diminishing the fear of cross-contamination and improving current practices.1
The use of GFP reporters offers an attractive method for easily distinguishing controls strains from true contamination. This reporter system is well characterized, highly stable, and has been used for a number of biological applications. Further, this reporter protein readily exhibits a fluorescent signal upon exposure to UV light and does not require the presence of co-factors or substrates for its use, allowing for a rapid, easy, non-invasive mechanism for visualizing bacteria.
In this study, GFP-labeled E. coli strains were created using the ATUM IP-Free Dasher GFP construct, which is a synthetic, non-Aequorea, plasmid-based expression system. Here, Shiga toxin-producing O157 and 4 of the “Big Six” non-O157 E. coli serogroups were developed as reporter-labeled positive controls (Table 1).
Table 1. ATCC reporter-labeled strains.
| ATCC number | Strain description | Serotype | Genotype |
|---|---|---|---|
| 35150GFP | Escherichia coli O157:H7 GFP | O157:H7 | stx1+, stx2+, eaeA+ |
| 51657GFP | Escherichia coli O157:H7 GFP | O157:H7 | stx1+, stx2+, eaeA+ |
| BAA-2196GFP | Escherichia coli O26:H11 GFP | O26:H11 | stx1+, stx2+, eaeA+ |
| BAA-2215GFP | Escherichia coli O103:H11 GFP | O103:H11 | stx1+, stx2-, eaeA+ |
| BAA-2209GFP | Escherichia coli O111 GFP | O111 | stx1+, stx2+, eaeA+ |
| BAA-2219GFP | Escherichia coli O121:H19 GFP | O121:H19 | stx1-, stx2+, eaeA+ |
Reporter signal detection
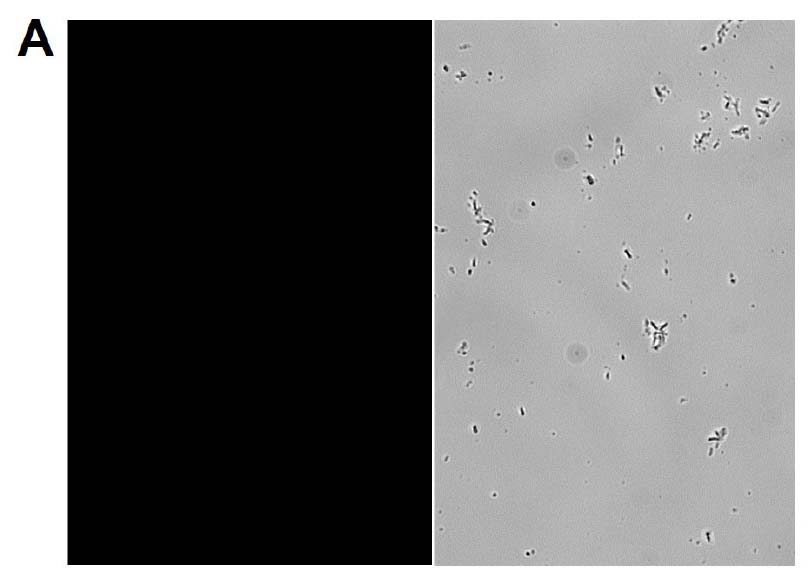
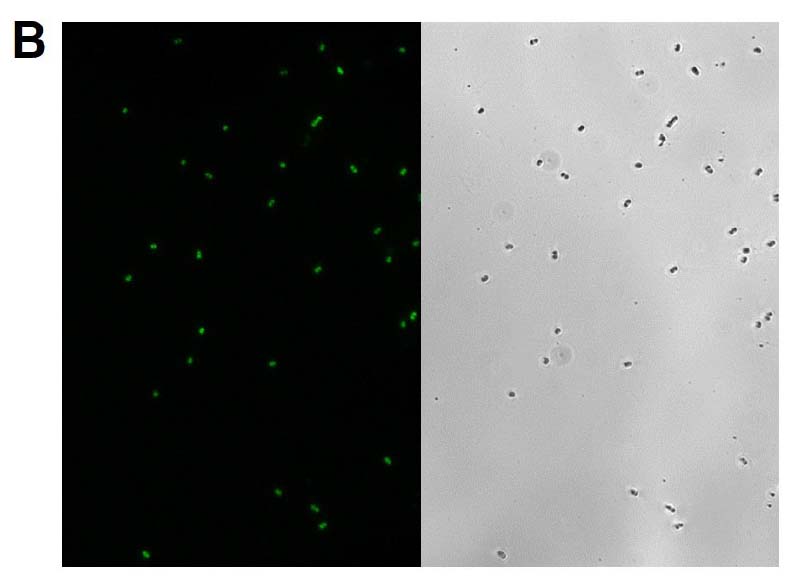
The GFP signal from reporter-labeled strains was detected by exposing colonies to UV light (Figure 1). The fluorescence from individual cells was also confirmed via microscopy and flow cytometry using the appropriate excitation and emission (Figure 2).
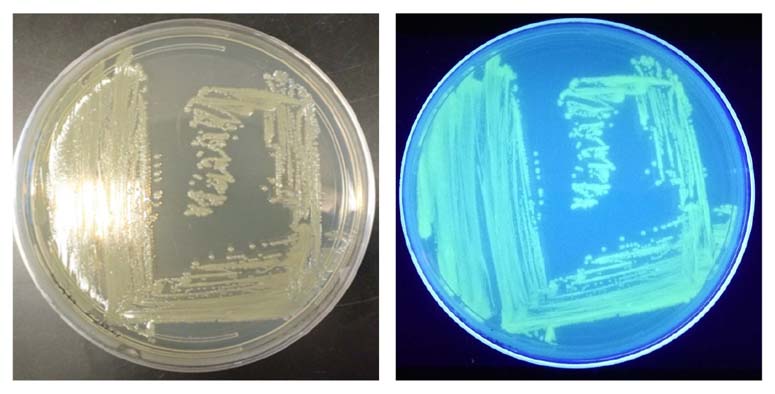
Figure 1. Visualization of the reporter-labeled strains. Toxinogenic ATCC BAA-2209 transformed with a plasmid bearing gfp (ATCC BAA-2209GFP) was grown at 37°C on Tryptic Soy Agar (TSA). Visualization was realized by exposing the ATCC BAA-2209GFP plate to UV light (302 nm).
 |
 |
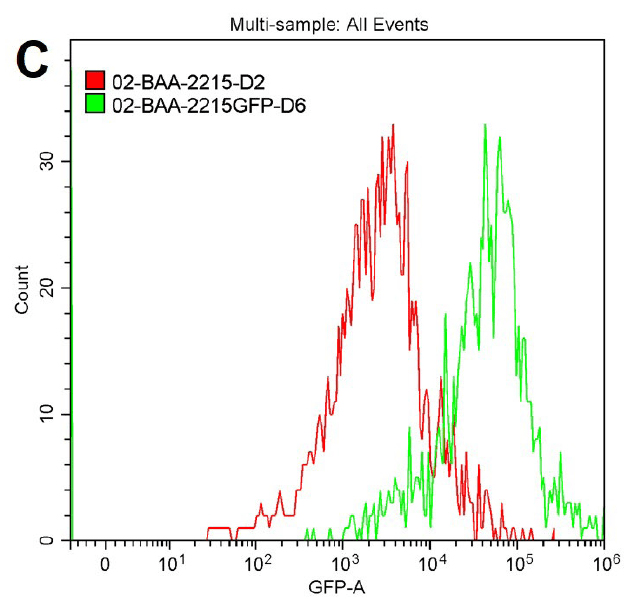 |
Figure 2. Microscopy and flow cytometry analyses from representative reporter-labeled and progenitor strains. Evaluation of cells through the use of a 40X objective with a 488 nm filter and phase contrast demonstrated that (A) progenitor ATCC BAA-2219 cells did not show any fluorescence, while (B) transformed ATCC BAA-2219GFP cells showed green fluorescence. GFP presence was also determined by flow cytometry analysis. (C) The overlaid histogram exhibits data from progenitor (red) and reporter-labeled E. coli cells (green) acquired on a CytoFLEX Cytometer (Beckman Coulter, Inc.) using a 488 nm laser and GFP filter set. The figure demonstrates a clear separation of the reporter-labeled GFP strain and the progenitor strain.
Chromogenic medium
Chromogenic media may be used to assist in the identification E. coli serotypes; the color of colonies from reporter-labeled strains and their progenitor strains should be in the same color family. GFP-labeled strains were compared with their progenitor strains to identify phenotypic changes on Rainbow Agar (Biolog). Color differences between reporter-labeled and progenitor strains were minimal (Figure 3).
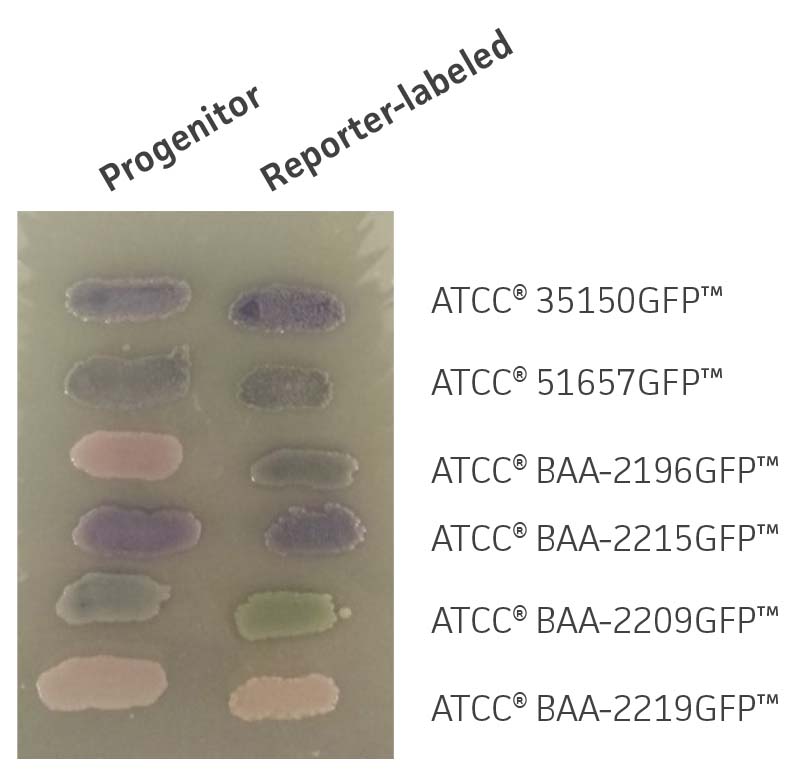
Figure 3. Chromogenic phenotype on rainbow agar. Rainbow Agar was prepared according to manufacturer specifications. The progenitor strains (left) and the reporter-labeled strains (right) were streaked on a plate and incubated for 16h at 37°C. The chromogenic properties of progenitor strains were compared to those of reporter-labeled strains.
Growth effects
To determine whether gfp affects the growth and viability of the reporter-labeled E. coli, growth studies were performed (Figure 4). The growth constant (k) varied from 2.3±0.5 to 3.6±1.2 generations/hour in the absence of the plasmid (progenitor) and from 1.6±1.1 to 1.9±0.9 generations/hour in the presence of the plasmid (reporter-labeled). The maximum decrease in the growth constant was a 45.8% drop for ATCC BAA-2219GFP and ATCC BAA-2196GFP. All changes were within an acceptable range as these strains were intended for qualitative assays that simply require visible growth after overnight culturing on plates.
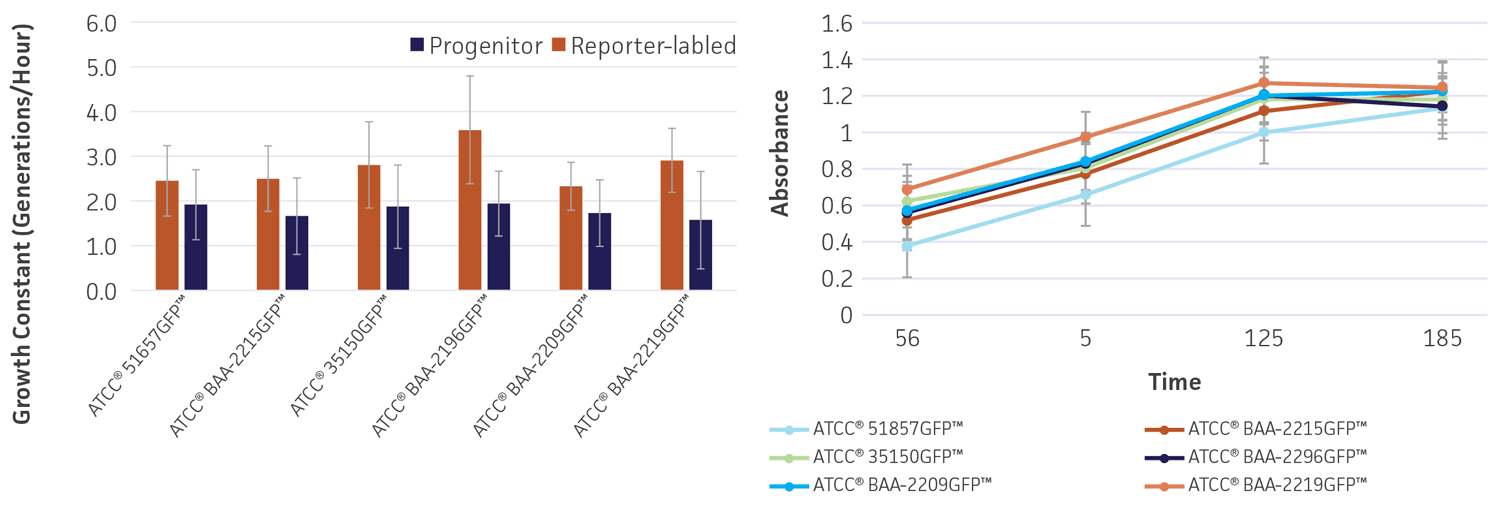
Figure 4. Growth rate of progenitor and reporter-labeled strains. Growth curves were performed in triplicate to determine the (A) growth constant (k) and (B) growth curve. Using the Bioscreen C MBR (Oy Growth Curves Ab Ltd.), 200 μL cultures were prepared with a 1:100 inoculum from an overnight culture and were incubated at 37°C in Tryptic Soy Broth (TSB) with constant shaking. Error bars represent standard error.
Plasmid stability
To determine the stability of the gfp plasmid, the engineered reporter-labeled strains were passaged once every 24 hours under temperature stress at 42°C to mimic the testing conditions used for STEC. The percentage of GFP positive colonies varied depending on the strain, ranging from 87-100% of the population after 2 days (Figure 5). This level of plasmid stability is within an acceptable range for the intended qualitative testing workflow.
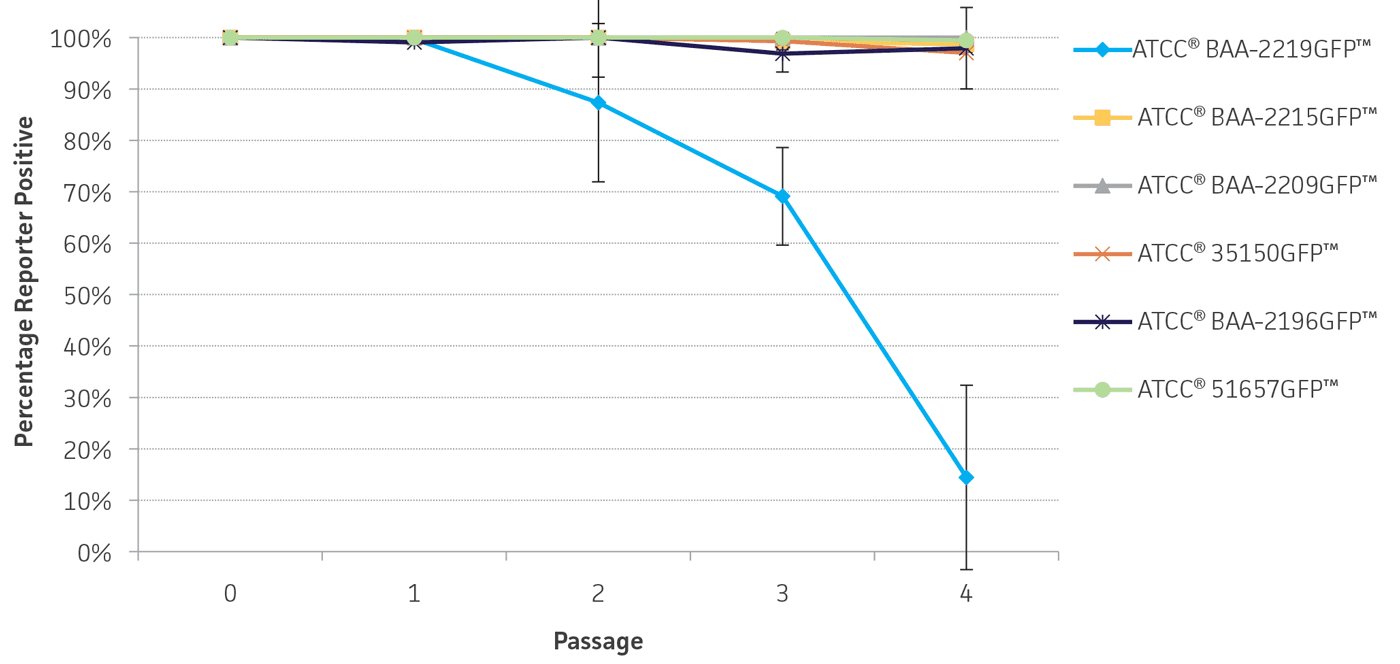
Figure 5. Plasmid stability. Reporter-labeled strains were grown in 5 mL of TSB at 42°C and passaged 1:100 into fresh TSB once every 24 hours. A serial dilution was performed to obtain a countable number of colonies. 100 μL of an appropriate dilution was plated on TSA and incubated overnight at 37°C. The percentage of colonies expressing the reporter was recorded daily over 4 days. Error bars represent standard deviation.
Conclusion
In this study, multiple serogroups of E. coli were engineered with GFP reporters. Phenotypic changes between the progenitor and reporter-labeled strains were minimal on chromogenic medium. As expected, growth rate differences between the progenitor and reporter-labeled strains were present in liquid culture but were acceptable for the qualitative assays for which the strains were designed. The gfp plasmid was stable in bacterial populations for ≥2 days. These reporter-labeled bacteria strongly emit light and can be detected immediately after exposure to UV light, eliminating uncertainty about cross-contamination. This study demonstrates that GFP-labeled QC strains can be routinely used as positive controls to increase reliability in food testing assays.
Download a PDF of this application note
Download NowReferences
- “Background on the FDA Food Safety Modernization Act (FSMA).” U.S. Food and Drug Administration. U.S. Department of Health and Human Services, 13 July 2015. http://www.fda.gov/food/guidanceregulation/fsma/ucm239907.htm. Accessed 7 July 2016.
