Authors: Stephen King, MS; Brian Chase, PhD; Afshin Sohrabi, PhD; Maria Mayda, PhD; Kurt Langenbach, PhD; Cara N. Wilder, PhD
Abstract
Novel molecular-based methods offer an effective and efficient means for the detection of mycoplasma contamination in cell cultures. However, for these methods to become widely accepted in research laboratories, they must demonstrate sensitivity, particularly with regard to the limit of detection. Here, we describe the use of quantitative mycoplasma DNA controls in the evaluation of two molecular-based detection systems.
Download a PDF of this application note
Download NowIntroduction
Mycoplasma contamination affects roughly 15-35% of continuous cell cultures, resulting in a number of deleterious effects including the induction of chromosomal abnormalities, the disruption of DNA and RNA synthesis, and the inhibition of both cell metabolism and growth rate.1-3 Historically, the recommended mycoplasma testing protocol for research laboratories included the use of traditional culture-based techniques and DNA staining with fluorochromes.4
Despite the successful use of these detection methods for many years, their limit of detection remains undefined. Moreover, the entire testing procedure is laborious, costly, and difficult to interpret. However, before an alternative method, such as a molecular-based detection system, can become widely accepted and implemented, this method must exhibit sensitivity with regard to the limit of detection.
To this end, ATCC has designed quantitative mycoplasma DNA controls for use in testing and calibration in ISO 17025 accredited laboratories, inclusivity/exclusivity testing, establishing limits of detection, verification or comparison of test methods, and other molecular applications. Here, we demonstrate the use of these products in evaluating the sensitivity of two different molecular-based detection systems.
Establishing the sensitivity of molecular-based detection systems
The limit of detection was established for two molecular-based detection systems comprising a quantitative mycoplasma DNA control (Acholeplasma laidlawii, Mycoplasma orale, or Mycoplasma hominis), a proprietary reaction mixture containing pre-evaluated 16S rRNA specific primers and probes, and either the Bio-Rad QX100 Droplet Digital PCR (ddPCR) or CFX96 Touch Real-Time PCR (QPCR) platform. For each control, predicted input quantities of 1, 10, 100, 1000, 10000, and 100000 were measured using both systems, and the observed sample copy number was plotted (Figure 1). Each assay was performed twice (N=2), and the individual input quantity for each sample was tested in triplicate (n=6). The total mean and standard deviation of the combined samples were calculated at a 95% confidence interval using JMP Software (Table 1).
At each of the predicted input copy numbers, the mean observed copy number for the combined samples fell within the 95% confidence interval. In particular, for predicted input copy numbers of 1 and 10, the mean observed copy number was statistically closer in value to the predicted copy number when using the QPCR platform. These results indicate that both molecular-based detection systems are able to sensitively detect and quantify mycoplasma DNA, with the QPCR platform exhibiting more accuracy with lower concentrations of genomic material.
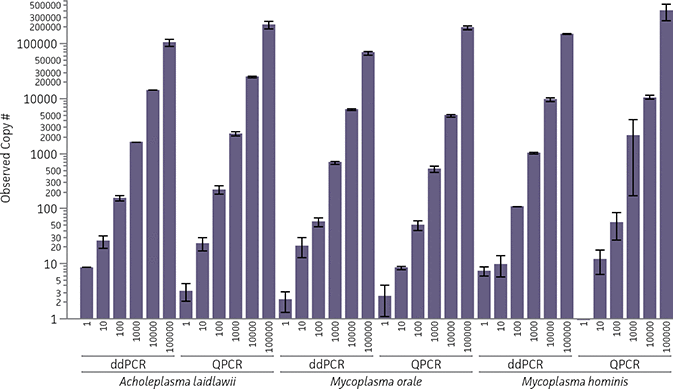
Figure 1. Observed sample copy number relative to predicted input copy number by ddPCR and QPCR for three quantitative mycoplasma DNA controls
Table 1. Observed total mean sample copy number relative to predicted input copy number measured by ddPCR and QPCR (N=2, n=6).
| Platform | Predicted copy number | Mean | Standard deviation | Confidence interval (95%) |
|---|---|---|---|---|
| ddPCR | 1 | 6.21 | 3.11 | 3.82 -8.61 |
| QPCR | 1 | 2.07 | 1.71 | 0.766 - 3.39 |
| ddPcR | 10 | 16.61 | 9.37 | 12.40 - 26.81 |
| QPCR | 10 | 15.16 | 8.36 | 8.74 - 21.59 |
| ddPCR | 100 | 110.78 | 44.51 | 76.57 - 144.99 |
| QPCR | 100 | 113.89 | 30.51 | 43.54 - 184.20 |
| ddPCR | 1000 | 1145.78 | 422.40 | 821.09 - 1470.05 |
| QPCR | 1000 | 1728.94 | 1367.80 | 677.56 - 2780.30 |
| ddPCR | 10,000 | 10408.90 | 3542.83 | 7685.60 - 13135 |
| QPCR | 10,000 | 13741.70 | 9094.59 | 6751 - 20732 |
| ddPCR | 100,000 | 111122 | 38150 | 81798 - 140447 |
| QPCR | 100,000 | 280089 | 119594 | 188161 - 372017 |
Conclusion
The use of quantitative external DNA controls provides an effective approach for the development and evaluation of novel molecular-based assays for the detection of mycoplasma contamination. Quantitative mycoplasma DNA controls are currently available as certified reference materials from ATCC.
Download a PDF of this application note
Download NowReferences
- Drexler H.G., and Uphoff C.C. Mycoplasma contamination of cell cultures: Incidence, sources, effects, detection, elimination, prevention. Cytotechnology 39, 75-90 (2002). PubMed: 3463982
- McGarrity G.J., Constantopoulos G., and Barranger J.A. Effect of mycoplasma infection on pyruvate dehydrogenase complex activity of normal and pyruvate dehydrogenase complex deficient fibroblasts. Exp Cell Res. 151, 557-562 (1984). PubMed: 2909916
- Rottern S. Interaction of Mycoplasma with Host Cells. Physiol Rev 83, 15 (2003). PubMed: 12663864
- Chandler D.K.F, Volokhov D.V., Chizhikov V.E. Historical overview of mycoplasma testing for production of biologics. Am Pharm Rev. (2011).
