Microbiome Research
Optimize your research with the right controls
The complexities involved in 16S rRNA community profiling and shotgun metagenomics methods pose significant challenges for microbiome research. Significant biases can be introduced at each stage of the microbiome workflow, affecting data interpretation and reproducibility.
NGS Standards provide a solution to this problem. From sample collection to data analysis, NGS Standards enable you to optimize your diverse research applications with confidence and improve the consistency and reproducibility of your data, run after run.
The robust applicability of these controls, combined with the ATCC commitment to authentication and characterization, make NGS Standards ideal tools for standardizing data from a wide range of sources and generating consensus among microbiome applications and analyses.
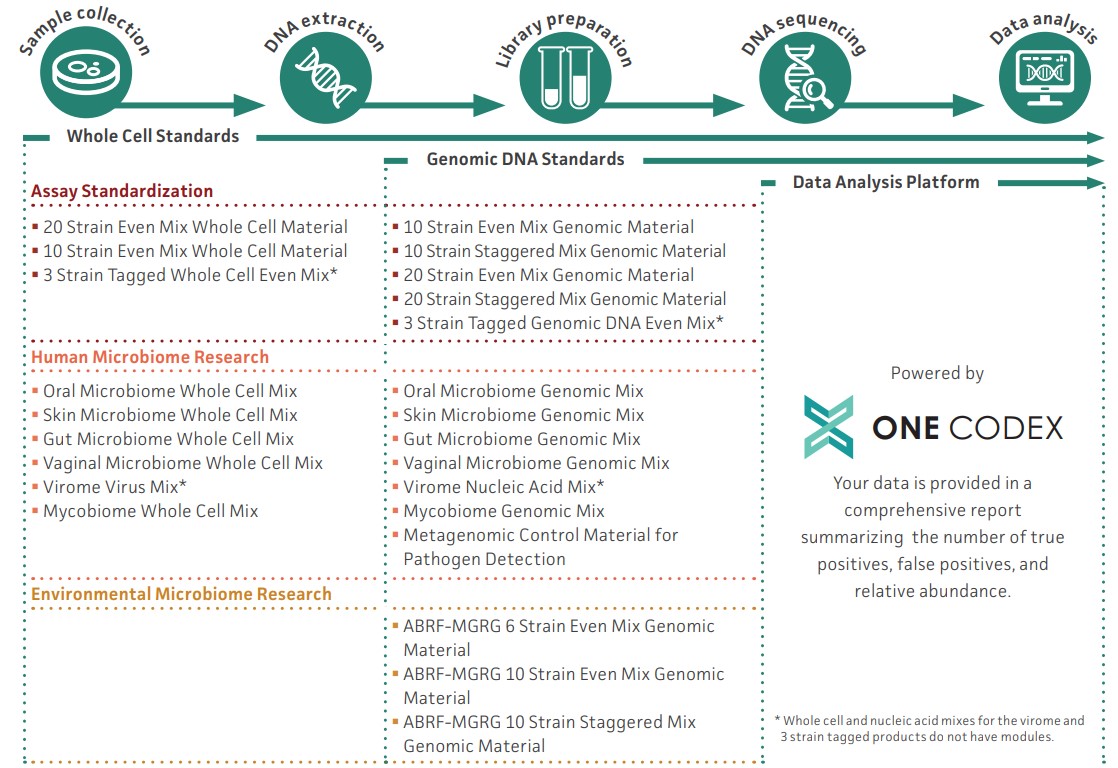
Microbiome research solutions

Data analysis tool
Bring your microbiome research to the next level by combining the power of physical laboratory standards with state-of-the-art bioinformatics.
Find Solutions
Application data
Check out our application data to see how the experts are using NGS Standards to improve reproducibility in microbiome research.
Get the DataImprove assay consistency
NGS Standards support a broad array of applications ranging from method optimization to data interpretation, and they serve as superior controls for microbial community testing and assay development on any platform. By using NGS Standards in your research, you can compare the performance and overall accuracy of your methods and ensure the validity of your results. Read our application note to discover how to incorporate these standards into your next project.
View the application note