Reliable diagnostic tools depend on credible reference materials
The highly pathogenic nature and transmission dynamics of SARS-CoV-2 have necessitated the availability of rapid, robust detection methods to ensure that infected individuals are treated in a timely manner. For SARS-CoV-2 molecular diagnostics manufacturers moving from emergency use authorization (EUA) to 510(k) premarket submission to the US Food & Drug Administration (FDA), robust testing must be performed to demonstrate that the device is safe and effective as well as substantially equivalent to legally marketed devices. To support these validation studies, ATCC provides an extensive array of authenticated and clinically relevant materials for evaluating limit of detection, inclusivity, and cross-reactivity.
Explore our resources for assay development
Molecular Diagnostic Tools
We provide a variety of ready-to-use standards that can be used throughout the assay development process. Explore our collection of molecular standards and inactivated organisms for molecular diagnostics development.
In Vitro Diagnostics
Health care providers rely on diagnostic tests to detect disease and monitor health. Discover why advanced biological models are essential for the development and validation of these essential tools.
Design your SARS-CoV-2 assay
Our extensive collection contains a variety of viral, bacterial, and fungal species for SARS-CoV-2 molecular diagnostics development, including SARS-CoV-2 reference materials, high-priority pathogens from the same genetic family as SARS-CoV-2, and high-priority organisms likely present in respiratory specimens. To support your unique assay development needs, we provide these materials in a variety of formats.
- Heat-inactivated preparations – Inviable, non-replicative preparations that are quantitated by Droplet Digital PCR or qPCR. These materials are ideal for use in molecular assays or as a process control (e.g., nucleic acid extraction through the qPCR amplification process).
- Live strains – Minimally passaged, fully characterized strains that are authenticated using a polyphasic approach that confirms genotype and phenotype.
- Genomic nucleic acids – Whole-genome preparations aseptically prepared from minimally passaged ATCC cultures. Each preparation is supported by stringent quality control testing to ensure product authenticity and functionality.
- Synthetic nucleic acids – Quantitative DNA and RNA synthetically manufactured under ISO 13485 guidance to include key target regions from select viral, bacterial, and protozoan species. These standards enable researchers to work with higher concentrations of target nucleic acid sequences for unculturable or high-containment species.
- Respiratory inclusivity and exclusivity panels – ATCC’s respiratory inclusivity and exclusivity panels are composed of authenticated nucleic acids or heat-inactivated material derived from respiratory pathogens commonly cited for use as control materials in SARS-CoV-2 EUA submissions and/or FDA 510(k) submissions.
ATCC resources for SARS-CoV-2 molecular diagnostic development:
| Organism | Heat-inactivated Strains | Live Strains | Genomic Nucleic Acids | Synthetic Nucleic Acids |
|---|---|---|---|---|
| SARS-CoV-2 | ✓ | ✓ | ✓ | |
| Human coronavirus 229E | ✓ | ✓ | ||
| Human coronavirus HKU1 | ✓ | |||
| Human coronavirus NL63 | ✓ | |||
| Human coronavirus OC43 | ✓ | ✓ | ||
| MERS-CoV | ✓ | |||
| SARS-CoV | ✓ |
Table 2. Viruses
| Organism | Heat-inactivated Strains | Live Strains | Genomic Nucleic Acids | Synthetic Nucleic Acids |
|---|---|---|---|---|
| Adenovirus 1 | ✓ | ✓ | ||
| Adenovirus 4 | ✓ | ✓ | ||
| Adenovirus 7 | ✓ | ✓ | ||
| Bocavirus | ✓ | |||
| Cytomegalovirus (HHV-5) | ✓ | ✓ | ||
| Enterovirus A (EV-A71) | ✓ | |||
| Enterovirus B (Echovirus 6) | ✓ | |||
| Enterovirus C (Coxsackievirus A17) | ✓ | |||
| Enterovirus D (68) | ✓ | ✓ | ||
| Epstein-Barr virus (HHV-4) | ✓ | ✓ | ||
| Human herpesvirus 1 | ✓ | ✓ | ||
| Human herpesvirus 2 | ✓ | ✓ | ||
| Human metapneumovirus | ✓ | |||
| Influenza A virus subtype H1 | ✓ | ✓ | ||
| Influenza A virus subtype H3 | ✓ | ✓ | ||
| Influenza B virus | ✓ | ✓ | ||
| Measles virus | ✓ | ✓ | ||
| Mumps virus | ✓ | ✓ | ||
| Parainfluenza virus 1 | ✓ | ✓ | ||
| Parainfluenza virus 2 | ✓ | ✓ | ||
| Parainfluenza virus 3 | ✓ | ✓ | ||
| Parainfluenza virus 4 | ✓ | ✓ | ||
| Respiratory syncytial virus | ✓ | ✓ | ||
| Rhinovirus | ✓ | ✓ |
Table 3. Bacteria
Table 4. Fungi
| Organism | Heat-inactivated Strains | Live Strains | Genomic Nucleic Acids | Synthetic Nucleic Acids |
|---|---|---|---|---|
| Aspergillus flavus | ✓ | ✓ | ||
| Aspergillus fumigatus |
✓ | ✓ | ||
| Candida albicans | ✓ | ✓ | ||
| Cryptococcus neoformans |
✓ | ✓ | ||
| Pneumocystis jirovecii |
✓ |
*A quantitative genomic format is not currently available for this product
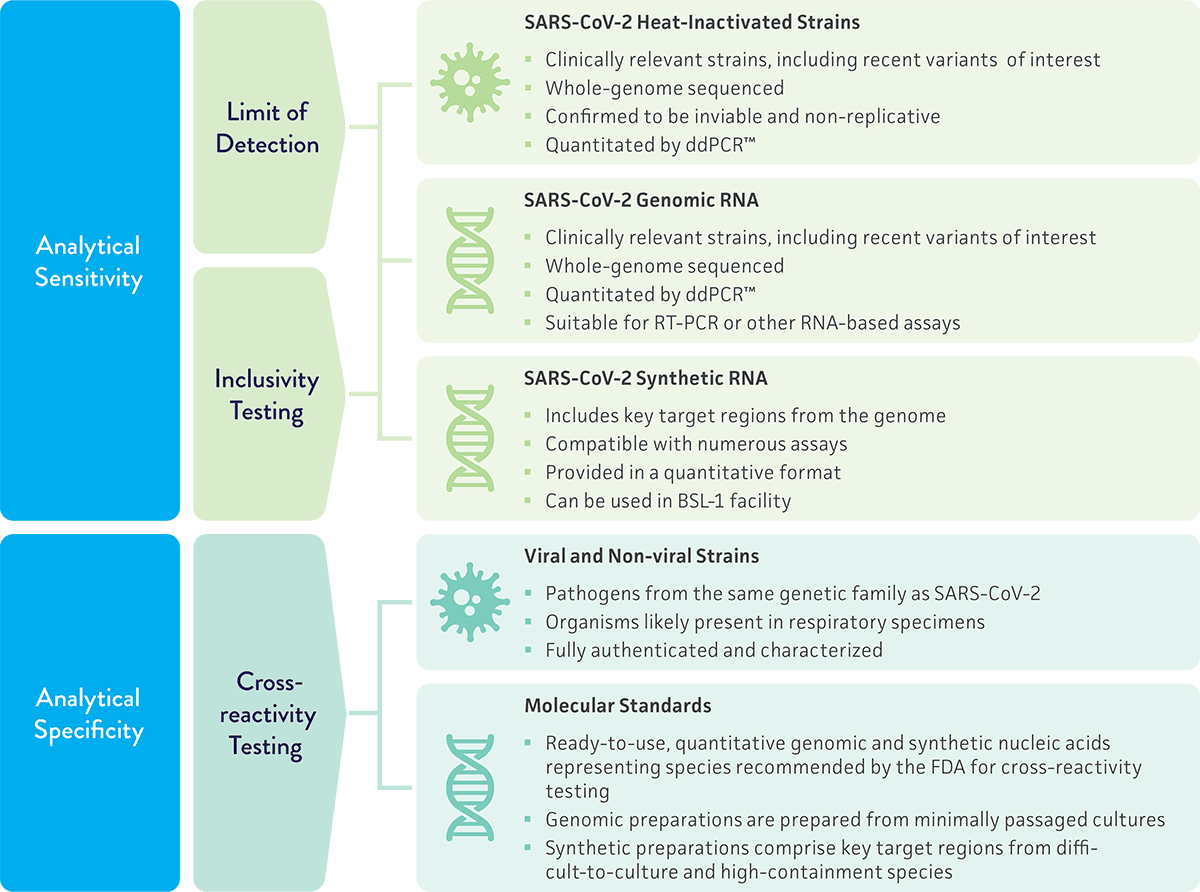
Evaluate assay performance
Because molecular diagnostic assays provide powerful tools for the rapid detection and identification of clinically relevant pathogens, it is essential that they are properly validated to guarantee accurate results and uncompromised data. To evaluate the performance of a novel molecular diagnostic test for SARS-CoV-2, we provide a variety of materials for validation studies on the limit of detection, inclusivity, and cross-reactivity of the assay. These studies help ensure that the assay is able to accurately detect SARS-CoV-2 without reacting to related pathogens.
- Limit of detection – The minimum amount of a target that can be accurately distinguished from the absence of a sample within a given level of confidence. For SARS-CoV-2 molecular diagnostic assays, the FDA recommends testing a quantitated inactivated viral preparation spiked into a clinical matrix as the inactivated preparation closely reflects the live virus in a clinical sample. Genomic RNA is also recommended as a material for testing.
- Inclusivity testing – Confirmation that your assay detects the intended target. Testing should include multiple strains of SARS-CoV-2 and demonstrate that the strains can be detected by the proposed assay. In silico analysis of published SARS-CoV-2 sequences is also acceptable.
- Cross-reactivity testing – Confirmation that your assay does not react with related pathogens or normal/pathogenic flora that may be present in the clinical sample. For this study, the FDA recommends testing high priority pathogens from the same genetic family and high priority organisms that are likely present in a respiratory specimen.
Watch our webinar to discover how to remove culturing from the equation
Synthetic SARS-CoV-2 RNA are compatible with existing assays
ATCC has developed four quantitative synthetic molecular standards for use as controls in the development of assays designed to detect and quantify SARS-CoV-2. Because these standards are synthetically derived, they eliminate the need to culture viruses in the laboratory and they are safe to use under biosafety level 1 conditions. These constructs are compatible with several assays, including the nucleocapsid-targeted real-time RT-PCR primer and probe sets developed by the CDC.
Table 2. ATCC synthetic SARS-CoV-2 RNA and compatible assays
| ATCC No. | Description | Compatible assays |
|---|---|---|
| VR-3276SD | Quantitative Synthetic SARS-CoV-2 RNA containing portions of ORF1ab, N, E, nsp12 (RdRp), and ORF1b-nsp14 genes |
China CDC Primers and probes for detection 2019-nCoV (24 January 2020) Diagnostic detection of Wuhan coronavirus 2019 by real-time RT-PCR – Charité, Berlin Germany (17 January 2020) Detection of 2019 novel coronavirus (2019-nCoV) in suspected human cases by RT-PCR – Hong Kong University (23 January 2020) PCR and sequencing protocol for 2019-nCoV - Department of Medical Sciences, Ministry of Public Health, Thailand (Updated 28 January 2020) US CDC Real-Time RT-PCR Panel for Detection 2019-Novel Coronavirus (28 January 2020) US CDC panel primer and probes– U.S. CDC, USA (28 January 2020) |
| VR-3277SD | Quantitative Synthetic SARS-CoV-2 RNA: containing a portion of Spike 5’ end gene. |
Detection of WN-Human1 sequence from clinical specimen. – National Institute of Infectious Diseases Japan (17 January 2020) |
| VR-3278SD | Quantitative Synthetic SARS-CoV-2 RNA: containing a portion of Spike 3’ end gene. |
PCR and sequencing protocols for 2019-nCoV- National Institute of Infectious Diseases Japan (24 January 2020) |
| VR-3279SD | Quantitative Synthetic SARS-CoV-2 RNA containing the portion of nsp9 and nsp12 (RdRp) genes |
RT-PCR assays for the detection of SARS-CoV-2 with RdRp - Institut Pasteur, Paris (2 March 2020) Diagnostic detection of Wuhan coronavirus 2019 by real-time RT-PCR – Charité, Berlin Germany (17 January 2020) |