
Authors: Helen Christina, MS, and Dev Mittar, PhD
Introduction
Development of the Molecular Standards
Generation of Standard Curves
Quantification of Native HBV and HCV
Conclusions
References
Abstract
ATCC has developed quantitated synthetic molecular standards for Hepatitis B virus (HBV) and Hepatitis C virus (HCV) for use as positive controls in molecular-based assays. These synthetic molecular standards comprise short conserved fragments representing the HBV and HCV genomes, respectively, and are compatible with numerous published assays. Moreover, they can be handled at biosafety level one (BSL-1), are stable, and can be used as quantitative controls in qPCR assays for the detection and quantification of these viruses.
Download a PDF of this application note
Download NowIntroduction
Viral hepatitis caused by HBV and HCV is a major health concern that affects millions of people worldwide. Patients are routinely monitored by quantitative reverse transcription PCR (qRT-PCR) for the presence of HCV RNA or by qPCR for HBV DNA in blood. Since these viruses are difficult to culture in vitro, obtaining control material for these molecular-based assays is a challenge. To address this problem, ATCC has developed quantitative synthetic molecular standards for HBV and HCV for use as controls in molecular assays used for the detection and quantification of these viruses from clinical samples.
In the following proof-of-concept study, we used the synthetic molecular standards for HBV and HCV to generate standard curves using published primer sets.1,2 Furthermore, we used the respective standard curves to quantify the 3rd WHO international standard for HBV and the 4th WHO international standard for HCV from the National Institute for Biological Standards and Control (NIBSC) using primer sets from published literature.1,2
Development of the molecular standards
The HBV and HCV molecular standards were developed under ISO 13485 by careful selection and synthesis of multigene constructs relevant to primers and probes commonly used to detect these viruses via PCR-based methods. The HBV preparation includes fragments from the polymerase, surface, and X regions, and the HCV preparation includes the 5´UTR and 3´UTR regions (Table 1). Both preparations were quantified by Droplet Digital PCR (ddPCR; Bio-Rad) to ensure precise copy number and stabilized using a proprietary stabilization matrix.
Table 1. ATCC Synthetic Molecular Standards
| ATCC number | Description | Preparation |
|---|---|---|
| VR-3232SD | Synthetic Hepatitis B Virus (HBV) DNA | Fragments from the precore, core, P, S, and X regions |
| VR-3233SD | Synthetic Hepatitis C Virus (HCV) RNA | Fragments from the 5’UTR and X-tail region (3’UTR) |
Generation of standard curves using the HBV and HCV molecular standards
Standard curves were generated using serial ten-fold dilutions of the respective synthetic molecular standard DNA or RNA, ranging from 3 copies to 3 × 105 copies/reaction for HBV (Figure 1) and from 10 copies to 1 × 105 copies/reaction for HCV (Figure 2). DNA or RNA samples and standards were tested in triplicate. Assays were performed using the CFX96 Real-Time PCR Detection System (Bio-Rad). The primer and probe sets from published qPCR or qRT-PCR assays1,2 were used under the following conditions. Cycling conditions for HBV primer sets were 50°C for 2 min, 95°C for 2 min, followed by 45 cycles of 95°C for 15 sec and 60°C for 30 sec. For HCV, primer set conditions were 50°C for 15 min and 95°C for 2 min, followed by 40 cycles of 95°C for 15 sec and 60°C for 30 sec. The relative fluorescence unit (RFU) baseline threshold was set automatically and genome copy numbers were calculated using CFX Manager 3.0 Software (Bio-Rad). From this analysis, it was determined that the HBV and HCV molecular standards are compatible with published primer sets over a broad linear dynamic range and exhibit minimal variability as evident from the slope and R2 values (Table 2).
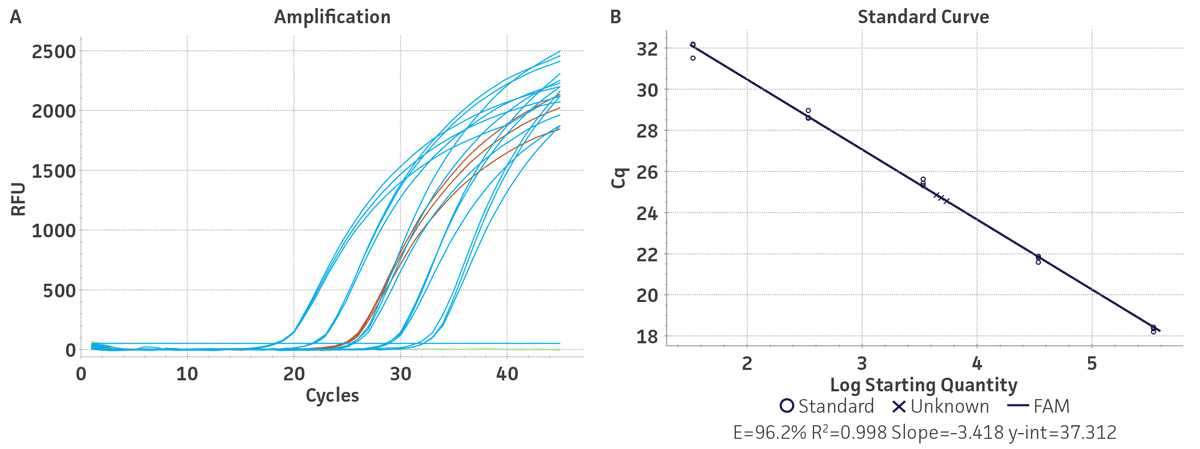
Figure 1. Generation of Standard Curves from Synthetic Molecular Standards for HBV. (A) Amplification plot and (B) standard curve generated using the HBV synthetic molecular standard (ATCC VR-3232SD) with the respective primer and probe set from the published Real-Time PCR Assay.1 (Blue) = Serial tenfold dilutions of the synthetic molecular standard. (Green) = Negative control.
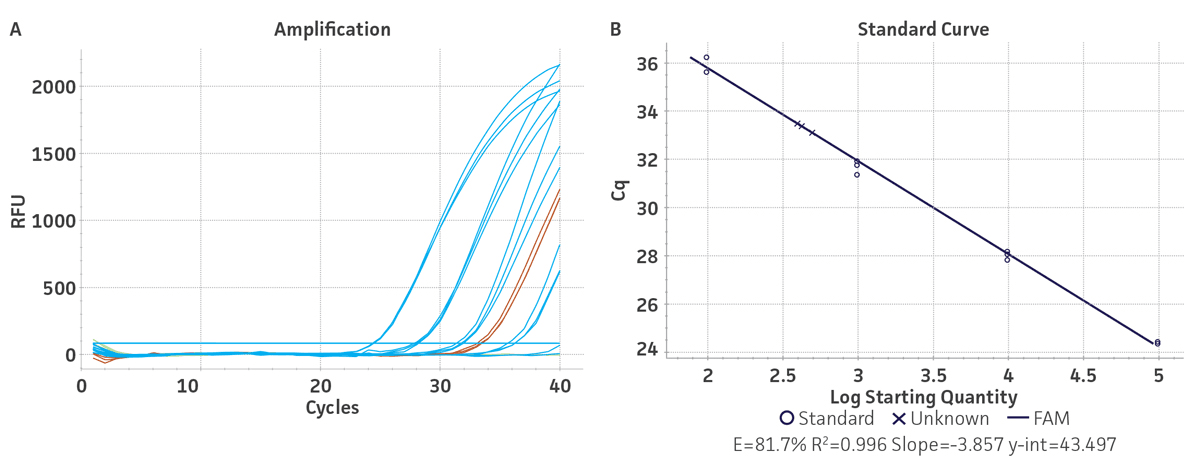
Table 2. Slope and R2 values generated using the HBV and HCV molecular standards.
| Molecular standard | Slope | R2 |
|---|---|---|
| HBV | -3.418 | 0.998 |
| HCV | -3.857 | 0.996 |
Quantification of native HBV and HCV nucleic acids using standard curves
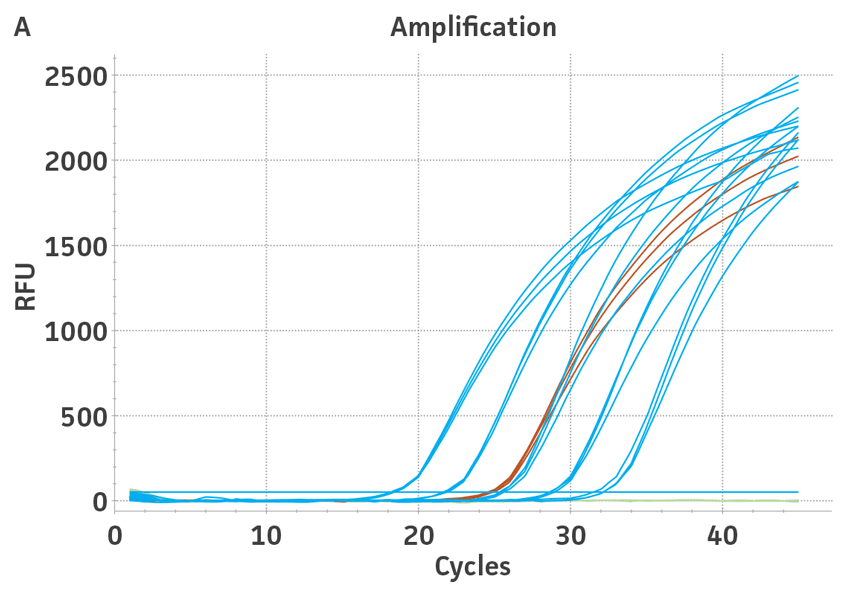
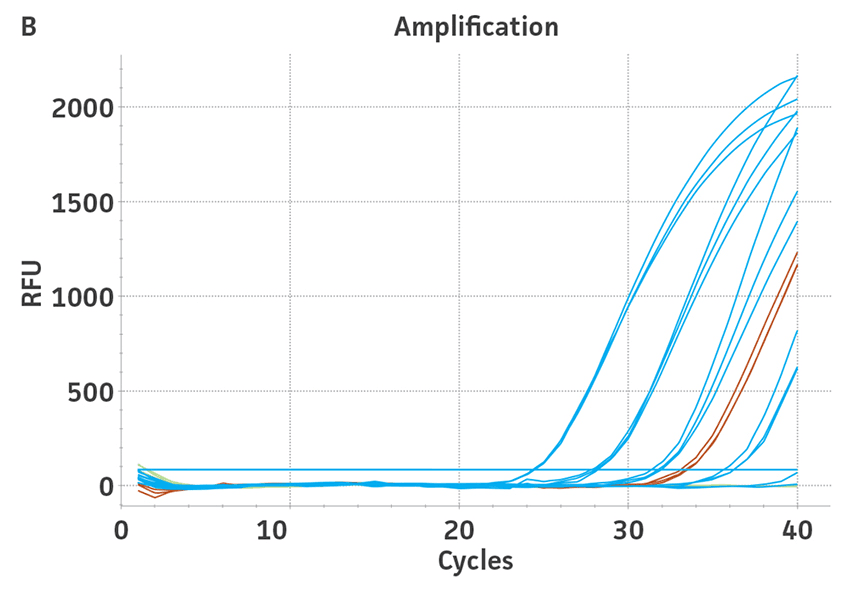
The 3rd WHO international standards for HBV (NIBSC code 10/264) and the 4th WHO international standards for HCV (NIBSC code 06/102) were obtained from NIBSC, UK. These standards contained 8.5 × 105 IU/mL and 2.6 × 105 IU/mL, respectively, as assigned by the WHO. Both standards were reconstituted in 0.5 mL of molecular grade water according to the manufacturer’s recommendations. Viral DNA or RNA from the respective WHO international standard was extracted using the QIAampR Viral RNA Mini Kit (QIAGENR). Each dilution was run in triplicate wells and repeated 3 times in independent experiments run on different days (n=9) (Figure 3).
Using the standard curve generated from the ATCC synthetic molecular standard for HBV and HCV under the published assay conditions,1,2 a value of 9.7 × 106 genome copies/mL was assigned to the 3rd WHO international standard and a value of 1.6 × 107 genome copies/mL was assigned to the 4th WHO international standard. By assigning a genome copy per mL value to WHO international standards, a conversion ratio of copies/mL to IU/mL can be easily calculated.3 In this study, using the specified qPCR and qRT-PCR conditions, we calculated that 1 IU/mL is equal to 11.4 and 61.5 genome copies/mL respectively of HBV and HCV synthetic molecular standards.
 |
 |
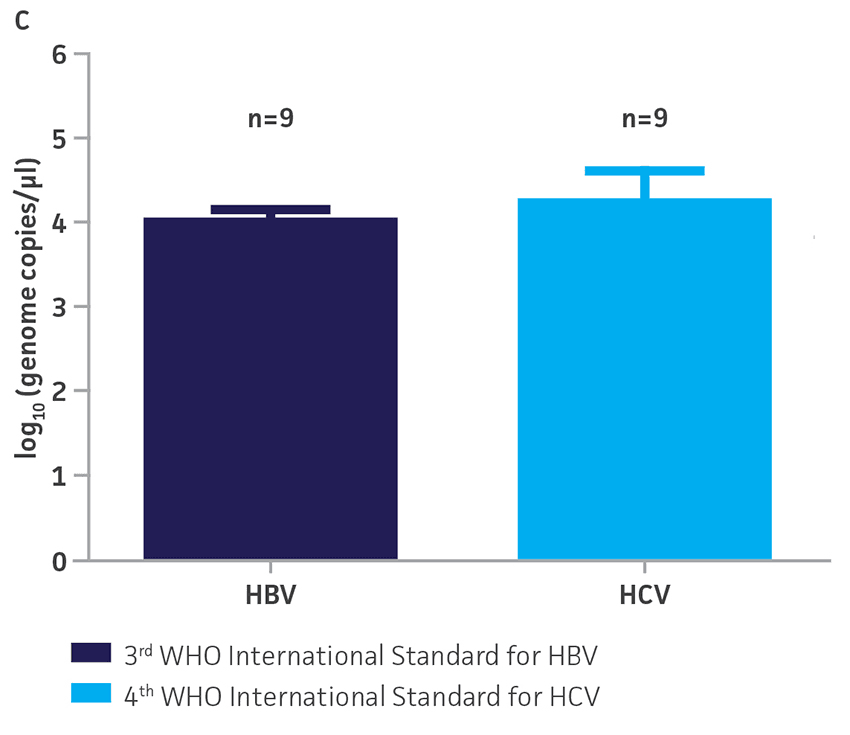 |
Figure 3. Quantification of WHO international standards for HBV and HCV using ATCC Synthetic Molecular Standards. An example of a qRT-PCR amplification plot showing the ATCC synthetic molecular standards (Blue) (A) HBV and the 3rd WHO international standard for HBV (Red). (B) HCV (Blue) and the 4th WHO international standard for HBV and negative control (Green). Both WHO international standards were diluted tenfold and run in triplicate wells in 3 independent experiments (n=9). (C) Quantitation of the WHO international standards for HBV and HCV samples respectively using the published assays for HBV1 and HCV.2 The average from triplicate wells from 3 independent experiments were used to calculate the quantities of HBV and HCV genomes respectively. Error bars indicate standard deviation and were calculated using GraphPad Prism software.
Conclusions
Here, we have generated synthetic molecular standards for HBV and HCV that can be used as positive controls in qPCR and qRT-PCR assays. These standards were generated under ISO 13485 and are quantitated, and stable, and can be handled in BSL-1 conditions. Through this study, we have demonstrated that both the HBV and HCV synthetic molecular standard are compatible with published assays and primer sets1,2 over a broad linear dynamic range, and exhibited minimal variability as evident from slope and R2 values. Moreover, the HBV and HCV standards can be used to determine the viral load of unknown samples through the generation of a standard curve. Taken together, these preparations provide a convenient, ready-to-use, well-characterized reference material for use in molecular-based assays.
Download a PDF of this application note
Download NowReferences
- Sun S, et al. Development of a new duplex real-time polymerase chain reaction assay for hepatitis B viral DNA detection. Virol. J. 8: 227, 2011.
- Lee SC, et al. Improved version 2.0 qualitative and quantitative AMPLICOR reverse transcription-PCR tests for hepatitis C virus RNA: calibration to international units, enhanced genotype reactivity, and performance characteristics. J. Clin. Microbiol. 38(11): 4171-4179, 2000.
- Welzel, et al. Real-time PCR assay for detection and quantification of Hepatitis B Virus Genotype A to G. J. Clin. Microbiol. 44(9):3325-3333. 2006.