
Authentication of a cell line is the sum of the processes by which a line’s identity is verified and shown to be free of contamination from other cell lines and microbes. Using standardized techniques, authentication enables communication among all users about the resource and ensures valid, reproducible experimental results. Without periodic testing, over-subcultured, misidentified, or cross-contaminated cell lines are released into the research arena resulting in spurious data.1 Clearly, the research community is better served by the use of tested material. However, guidelines for general testing of cell lines currently do not exist—especially for lines that are not obtained from a cell bank or are derived locally. In this technical document, ATCC recommends basic benchmark verification tests that can be employed by any lab and included in publication. We seek to engage the research community in an ongoing dialogue about this topic, including suggestions and ideas about the tests described in the technical document.
As a biological resource center, ATCC comprehensively performs authentication and quality-control tests on all distribution lots of cell lines. Therefore, obtaining and using low-passage cell lines from ATCC is a sure way to work and publish with confidence. When publishing research performed with an ATCC cell line, the designation, ATCC catalog number, and passage numbers under which experiments were conducted should be included in the materials and methods section of the research, for example NIH/3T3 (ATCC CRL-1658), passage number XX–YY.
ATCC recognizes that many cell lines used in basic and biomedical research are not available from ATCC or another cell bank. In some cases, these lines have undergone little or no authentication testing. In other cases, only outdated testing has been conducted. When performing research with these cell lines, it is good cell culture practice to conduct fundamental tests to ensure that the lines are not contaminated and correctly identified. When research with this material is published, details of the tests can and should be submitted to the journal editor and included in the materials and methods section of a manuscript.
The importance of passage number
Passage number is generally the number of times cells have been subcultured into a new vessel, usually within one lab. Avoiding the use of cell lines that have been in culture too long is an important step to ensure reliable and reproducible results. Unlike counting rings in a tree cross-section to determine age, passage number is not a property that can be tested or counted with a straightforward method. However, the consequences on experiments of using oversubcultured or high-passage cell lines remain. It is well documented that cell characteristics can change when cell lines are excessively subcultured.8-12 Cell lines that have been excessively subcultured can experience phenotypic changes as well as genotypic changes (genetic drift). It is also true that stocks of commonly used cell lines maintained in many laboratories have been passaged hundreds of times and should not be considered true models of the original source material.9,12,13
It is good cell culture practice to start experiments with fresh, low-passage cells and to use the cells in a predetermined range of passage numbers for best results. If you start to experience sudden and inexplicable variations in your experimental results, it may be that the cell line has been subcultured too often and needs to be replaced. By recording passage number, monitoring your cells with morphology checks, establishing and observing changes unique to the line you work with, you can help to promote the integrity of the cell banks which you have created in your research institution.
Recommended tests
Morphology check by microscope
Cellular morphology refers to the optical observation of a magnified cell culture. This can be the simplest and most direct method used to identify the state of cells. Obtaining morphology information from comparative observations both at high- and low-culture densities depends on knowledge of several factors. Morphology can vary between lines depending on the health of the cells and, in some cases, the differentiation state. Morphology can change with plating density as well as with different media and sera combinations. Cell morphology is best monitored through frequent, brief observations. In general, if a culture has an unusual appearance, there is likely a problem. It is recommended that researchers be alert during periodic morphology checks and maintain cell morphology images for comparisons.
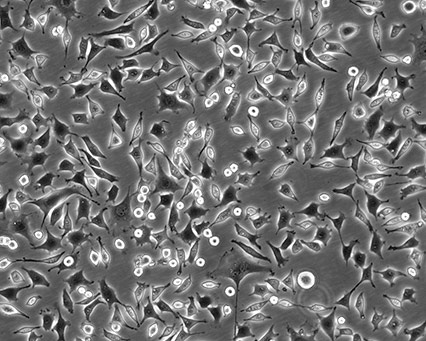 |
 |
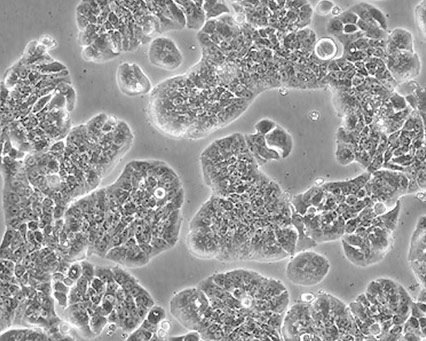 |
Figure 1. Three different ATCC cell lines at high densities.
Growth curve analysis
Evaluation of cell proliferation can yield valuable information about a culture’s response to a stimulus. Variable growth or sudden decreases or increases are signs that something may be amiss with your cell lines. Establishing baselines and quantifying cell culture growth are crucial elements for monitoring the consistency of the culture and determining a number of other factors, such as the best time to subculture, optimum dilution, and estimated plating efficiency at various cell densities. Growth curve analysis can also help determine population doubling times and should be performed routinely when enzymatic or functional analysis is imminent. As a rule, use cell lines with consistent growth properties.
Species verification by Isoenzymology
Isoenzyme analysis is used to verify the species of origin. Isoenzyme specimens are differentiated based on electrophoretic properties. Distribution patterns of a group of enzymes are characteristic of a particular species which simultaneously confirms the species identity and reveals contamination by another line of different species. A protocol may be found in Culture of Animal Cells: A Manual of Basic Technique by R. Ian Freshney, 5th ed. Pp. 275 – 278.
Identity verification with STR analysis (DNA fingerprinting) for human cell lines
DNA fingerprinting is a powerful tool in determining the identity and uniqueness of a human line. Short tandem repeat (STR) profiling establishes a DNA fingerprint for every human cell line and may be used as a record of the line. STR profiling, as performed by ATCC, uses multiplex PCR to simultaneously amplify the amelogenin gene and seventeen polymorphic markers, including the most informative polymorphic markers in the human genome. The pattern of repeats results in a unique STR identity profile for each cell line analyzed. The profile can be used as a baseline for comparison with future tests. ATCC uses the Promega PowerPlex 18D system and the ThermoFisher Scientific GeneMapper ID-X v1.2 software for analysis of the amplicons.
Mycoplasma detection
Mycoplasma contamination, a major problem in cell culture, can have adverse effects on cell lines, altering cell behavior and metabolism in many ways.2-7 Periodic assays to detect mycoplasma are critical for all continuous cell lines. A relatively easy and reliable biochemical method for detecting mycoplasma in a cell culture is to use Hoechst 33258, a fluorescent dye that binds specifically to DNA. Fluorescent Hoechst staining reveals mycoplasma contamination through their characteristic patterns of extracellular particulate or filamentous fluorescence at 500X magnification.
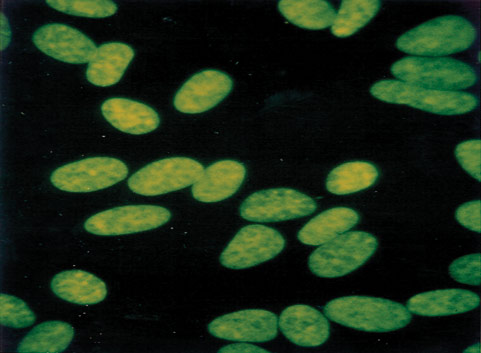
Figure 2. A cell line negative for mycoplasma.
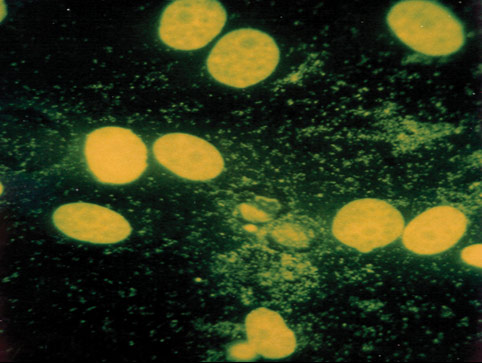
Figure 3. A cell line contaminated with mycoplasma.
Conclusion
While tests are always a good idea, your intuitions should not be ignored. When in doubt about the state of your cell lines, start with a new vial from your cell bank. By treating your cell lines as standard research components and providing verification benchmarks in publication, the research community is better served and information exchange between colleagues is enhanced. The result will be more reliable and reproducible experimental data.
When publishing with a cell line from ATCC, always reference the line with the common name followed by the ATCC catalog number. For example, cite as "NIH/3T3 (ATCC CRL-1658)" in your publications.
Download a PDF of this technical document
Download NowReferences
- Chatterjee R. Cases of Mistaken Identity. Science. 315:928-931, 2007.
- Drexler HG, et al. Mix-ups and mycoplasma: the enemies within. Leuk Res 26(4): 329 333, 2002.
- Kagemann G, et al. Impact of Mycoplasma hyorhinus infection of L-arginine metabolism: differential regulation of the human and murine iNOS gene. Biol Chem 386(10): 1055-1063, 2005.
- Lincoln CK, et al. Cell culture contamination: sources, consequences, prevention and elimination. Methods Cell Biol 57: 49-65, 1998.
- McGarrity GJ, et al. Cell culture mycoplasmas. In: The Mycoplasma, Vol. IV. Razin S and Barile MF, eds. New York: Academic Press. 353-390, 1985.
- Mirjalili A, et al. Microbial contamination of cell cultures: a two-year study. Biologicals 33(2): 81-85, 2005.
- No authors listed. Contamination of cell lines—a conspiracy of silence. Lancet Oncol 2(7): 393, 2001.
- Esquenet M, et al. LNCaP prostatic adenocarcinoma cells derived from low and high passage numbers display divergent responses not only to androgens but also to retinoids. J Steroid Biochem Mol. Biol 62(5-6): 391-399, 1997.
- Briske-Anderson MJ, et al. Influence of culture time and passage number on the morphological and physiological development of Caco-2 cells. Proc Soc Exp Biol Med 214(3): 248-257, 1997.
- Yu H, et al. Evidence for diminished functional expression of intestinal transporters in Caco-2 cell monolayers at high passages. Pharm Res 14(6): 757-762, 1997.
- Sambuy Y, et al. The Caco-2 cell line as a model of the intestinal barrier: Influence of cell- and culture-related factors on Caco-2 functional characteristics. Cell Biol Toxicol 21(1): 1-26, 2005.
- Wenger SL, et al. Comparison of established cell lines at different passages by karyotype and comparative genomic hybridization. Biosci Rep 24(6): 631-639, 2004.
- MacLeod RA, et al. Identity of original and late passage Dami megakaryocytes with HEL erythroleukemia cells shown by combined cytogenetics and DNA fingerprinting. Leukemia 11(12): 2032-2038, 1997.
